
Mongoose feces-associated gemycircularvirus c
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus humas3; Human associated gemykibivirus 3
Average proteome isoelectric point is 7.95
Get precalculated fractions of proteins
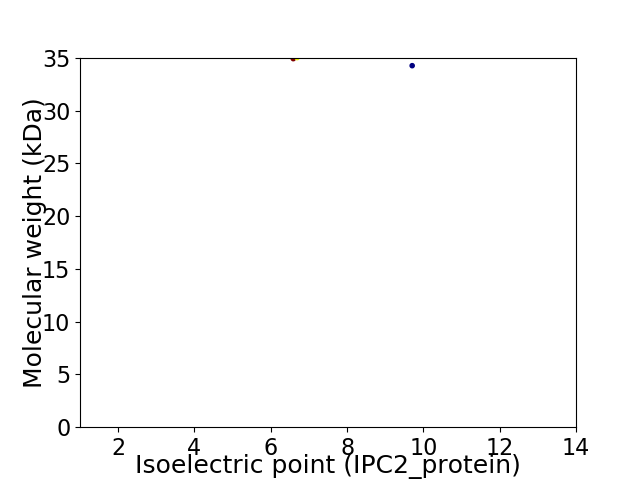
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E3JHI1|A0A0E3JHI1_9VIRU Replication-associated protein OS=Mongoose feces-associated gemycircularvirus c OX=1634488 PE=3 SV=1
MM1 pKa = 7.83SSFRR5 pKa = 11.84FQARR9 pKa = 11.84YY10 pKa = 10.11GLFTYY15 pKa = 7.62SQCGQLRR22 pKa = 11.84PSAVVEE28 pKa = 4.27LFSSLSADD36 pKa = 3.53CIVGRR41 pKa = 11.84EE42 pKa = 3.97AHH44 pKa = 6.62GDD46 pKa = 3.59GGTHH50 pKa = 6.01LHH52 pKa = 6.52AFVDD56 pKa = 4.76FRR58 pKa = 11.84RR59 pKa = 11.84KK60 pKa = 9.82YY61 pKa = 8.38RR62 pKa = 11.84TRR64 pKa = 11.84DD65 pKa = 3.08VRR67 pKa = 11.84KK68 pKa = 8.76FDD70 pKa = 3.82VEE72 pKa = 4.29GFHH75 pKa = 7.04PNIISTIRR83 pKa = 11.84SPSGSWDD90 pKa = 3.46YY91 pKa = 10.31ATKK94 pKa = 10.88DD95 pKa = 3.14GDD97 pKa = 3.76VCGGSLRR104 pKa = 11.84RR105 pKa = 11.84PEE107 pKa = 4.3PSQGSNAEE115 pKa = 4.02RR116 pKa = 11.84QNAGWDD122 pKa = 3.55QLRR125 pKa = 11.84DD126 pKa = 3.16ASTRR130 pKa = 11.84EE131 pKa = 3.56EE132 pKa = 4.16FLNLRR137 pKa = 11.84QRR139 pKa = 11.84IFSAISSNHH148 pKa = 5.62SLHH151 pKa = 6.82SLHH154 pKa = 5.94TQIGSLEE161 pKa = 4.38WIDD164 pKa = 3.88HH165 pKa = 6.68LIVTLANTRR174 pKa = 11.84SILHH178 pKa = 5.87NMVTFGNGLGDD189 pKa = 3.53IYY191 pKa = 11.45VWGPSRR197 pKa = 11.84TGKK200 pKa = 8.59TMWARR205 pKa = 11.84SLGKK209 pKa = 9.22HH210 pKa = 5.95AYY212 pKa = 9.85FGGLFSLEE220 pKa = 4.68EE221 pKa = 4.14YY222 pKa = 10.35EE223 pKa = 4.64QDD225 pKa = 3.58PDD227 pKa = 3.07CSYY230 pKa = 11.93AVFDD234 pKa = 5.63DD235 pKa = 3.64IQGGFKK241 pKa = 10.46FFPGYY246 pKa = 10.3KK247 pKa = 9.24SWLGQQTEE255 pKa = 5.26FYY257 pKa = 9.95CTDD260 pKa = 3.35RR261 pKa = 11.84YY262 pKa = 10.36RR263 pKa = 11.84KK264 pKa = 9.29KK265 pKa = 10.77KK266 pKa = 9.59HH267 pKa = 5.74IAWGKK272 pKa = 7.54PCIWLSNNNPMGEE285 pKa = 4.28DD286 pKa = 3.9ADD288 pKa = 4.7HH289 pKa = 7.46DD290 pKa = 4.16WLEE293 pKa = 4.16KK294 pKa = 10.65NCIIVYY300 pKa = 10.33VGDD303 pKa = 3.49RR304 pKa = 11.84LYY306 pKa = 11.41
MM1 pKa = 7.83SSFRR5 pKa = 11.84FQARR9 pKa = 11.84YY10 pKa = 10.11GLFTYY15 pKa = 7.62SQCGQLRR22 pKa = 11.84PSAVVEE28 pKa = 4.27LFSSLSADD36 pKa = 3.53CIVGRR41 pKa = 11.84EE42 pKa = 3.97AHH44 pKa = 6.62GDD46 pKa = 3.59GGTHH50 pKa = 6.01LHH52 pKa = 6.52AFVDD56 pKa = 4.76FRR58 pKa = 11.84RR59 pKa = 11.84KK60 pKa = 9.82YY61 pKa = 8.38RR62 pKa = 11.84TRR64 pKa = 11.84DD65 pKa = 3.08VRR67 pKa = 11.84KK68 pKa = 8.76FDD70 pKa = 3.82VEE72 pKa = 4.29GFHH75 pKa = 7.04PNIISTIRR83 pKa = 11.84SPSGSWDD90 pKa = 3.46YY91 pKa = 10.31ATKK94 pKa = 10.88DD95 pKa = 3.14GDD97 pKa = 3.76VCGGSLRR104 pKa = 11.84RR105 pKa = 11.84PEE107 pKa = 4.3PSQGSNAEE115 pKa = 4.02RR116 pKa = 11.84QNAGWDD122 pKa = 3.55QLRR125 pKa = 11.84DD126 pKa = 3.16ASTRR130 pKa = 11.84EE131 pKa = 3.56EE132 pKa = 4.16FLNLRR137 pKa = 11.84QRR139 pKa = 11.84IFSAISSNHH148 pKa = 5.62SLHH151 pKa = 6.82SLHH154 pKa = 5.94TQIGSLEE161 pKa = 4.38WIDD164 pKa = 3.88HH165 pKa = 6.68LIVTLANTRR174 pKa = 11.84SILHH178 pKa = 5.87NMVTFGNGLGDD189 pKa = 3.53IYY191 pKa = 11.45VWGPSRR197 pKa = 11.84TGKK200 pKa = 8.59TMWARR205 pKa = 11.84SLGKK209 pKa = 9.22HH210 pKa = 5.95AYY212 pKa = 9.85FGGLFSLEE220 pKa = 4.68EE221 pKa = 4.14YY222 pKa = 10.35EE223 pKa = 4.64QDD225 pKa = 3.58PDD227 pKa = 3.07CSYY230 pKa = 11.93AVFDD234 pKa = 5.63DD235 pKa = 3.64IQGGFKK241 pKa = 10.46FFPGYY246 pKa = 10.3KK247 pKa = 9.24SWLGQQTEE255 pKa = 5.26FYY257 pKa = 9.95CTDD260 pKa = 3.35RR261 pKa = 11.84YY262 pKa = 10.36RR263 pKa = 11.84KK264 pKa = 9.29KK265 pKa = 10.77KK266 pKa = 9.59HH267 pKa = 5.74IAWGKK272 pKa = 7.54PCIWLSNNNPMGEE285 pKa = 4.28DD286 pKa = 3.9ADD288 pKa = 4.7HH289 pKa = 7.46DD290 pKa = 4.16WLEE293 pKa = 4.16KK294 pKa = 10.65NCIIVYY300 pKa = 10.33VGDD303 pKa = 3.49RR304 pKa = 11.84LYY306 pKa = 11.41
Molecular weight: 34.94 kDa
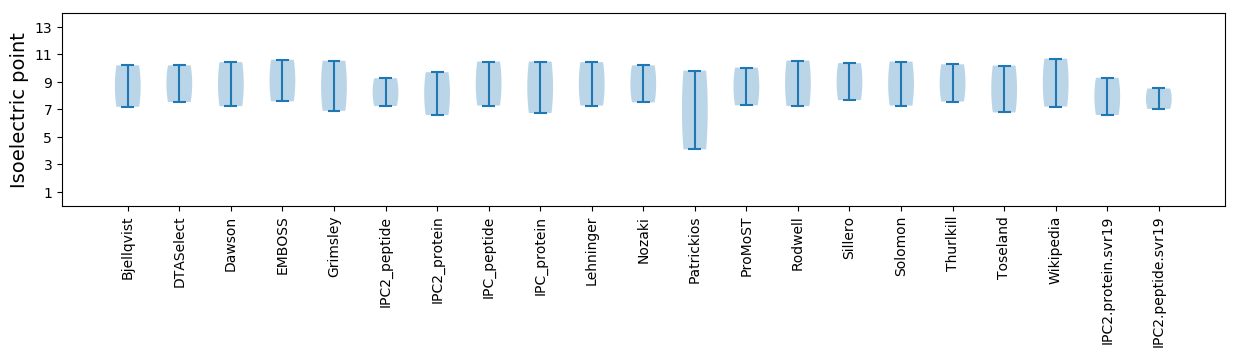
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E3JHI1|A0A0E3JHI1_9VIRU Replication-associated protein OS=Mongoose feces-associated gemycircularvirus c OX=1634488 PE=3 SV=1
MM1 pKa = 7.72AYY3 pKa = 10.24ARR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84FRR9 pKa = 11.84KK10 pKa = 7.9RR11 pKa = 11.84TYY13 pKa = 10.19ARR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84YY18 pKa = 6.01PTSRR22 pKa = 11.84RR23 pKa = 11.84KK24 pKa = 9.45PSRR27 pKa = 11.84RR28 pKa = 11.84SYY30 pKa = 9.57GRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SSGRR39 pKa = 11.84SVRR42 pKa = 11.84SIRR45 pKa = 11.84NIAARR50 pKa = 11.84KK51 pKa = 9.51CRR53 pKa = 11.84DD54 pKa = 2.87NMMGVPVSDD63 pKa = 3.91TNIAGPPAPVQLTAGTDD80 pKa = 3.45YY81 pKa = 11.62GFLFCPSARR90 pKa = 11.84RR91 pKa = 11.84VGSLDD96 pKa = 2.88GSLPRR101 pKa = 11.84SIHH104 pKa = 5.9ASASEE109 pKa = 3.93RR110 pKa = 11.84WKK112 pKa = 8.76TRR114 pKa = 11.84CYY116 pKa = 10.5IKK118 pKa = 10.68GLQEE122 pKa = 4.18TIEE125 pKa = 4.05VQPTDD130 pKa = 2.94GMGWLWRR137 pKa = 11.84RR138 pKa = 11.84IVFSCIGLLDD148 pKa = 4.05EE149 pKa = 4.99FPGVSVSVADD159 pKa = 3.68NVNGYY164 pKa = 9.09GRR166 pKa = 11.84ALWNVRR172 pKa = 11.84SGTAEE177 pKa = 3.85AQPLFTEE184 pKa = 4.38TANYY188 pKa = 9.96LFEE191 pKa = 4.28GTRR194 pKa = 11.84FRR196 pKa = 11.84DD197 pKa = 3.21WVNPFTAKK205 pKa = 10.34LDD207 pKa = 3.75RR208 pKa = 11.84SIVTVHH214 pKa = 7.01ADD216 pKa = 3.38TVRR219 pKa = 11.84TVQSNNPNGTYY230 pKa = 10.2KK231 pKa = 10.39VYY233 pKa = 10.75KK234 pKa = 9.7RR235 pKa = 11.84YY236 pKa = 10.34YY237 pKa = 8.05PINRR241 pKa = 11.84GIVYY245 pKa = 10.49ADD247 pKa = 4.05DD248 pKa = 3.63EE249 pKa = 4.81SGAGKK254 pKa = 10.6KK255 pKa = 9.98NDD257 pKa = 4.2AIYY260 pKa = 10.51AAGTTKK266 pKa = 10.7GNSGDD271 pKa = 4.48LFVLDD276 pKa = 4.06MFSAVGDD283 pKa = 4.07NEE285 pKa = 4.4GSTLNFGVHH294 pKa = 6.13ARR296 pKa = 11.84YY297 pKa = 8.98YY298 pKa = 7.44WHH300 pKa = 7.27EE301 pKa = 4.21GSGQQ305 pKa = 3.18
MM1 pKa = 7.72AYY3 pKa = 10.24ARR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84FRR9 pKa = 11.84KK10 pKa = 7.9RR11 pKa = 11.84TYY13 pKa = 10.19ARR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84YY18 pKa = 6.01PTSRR22 pKa = 11.84RR23 pKa = 11.84KK24 pKa = 9.45PSRR27 pKa = 11.84RR28 pKa = 11.84SYY30 pKa = 9.57GRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SSGRR39 pKa = 11.84SVRR42 pKa = 11.84SIRR45 pKa = 11.84NIAARR50 pKa = 11.84KK51 pKa = 9.51CRR53 pKa = 11.84DD54 pKa = 2.87NMMGVPVSDD63 pKa = 3.91TNIAGPPAPVQLTAGTDD80 pKa = 3.45YY81 pKa = 11.62GFLFCPSARR90 pKa = 11.84RR91 pKa = 11.84VGSLDD96 pKa = 2.88GSLPRR101 pKa = 11.84SIHH104 pKa = 5.9ASASEE109 pKa = 3.93RR110 pKa = 11.84WKK112 pKa = 8.76TRR114 pKa = 11.84CYY116 pKa = 10.5IKK118 pKa = 10.68GLQEE122 pKa = 4.18TIEE125 pKa = 4.05VQPTDD130 pKa = 2.94GMGWLWRR137 pKa = 11.84RR138 pKa = 11.84IVFSCIGLLDD148 pKa = 4.05EE149 pKa = 4.99FPGVSVSVADD159 pKa = 3.68NVNGYY164 pKa = 9.09GRR166 pKa = 11.84ALWNVRR172 pKa = 11.84SGTAEE177 pKa = 3.85AQPLFTEE184 pKa = 4.38TANYY188 pKa = 9.96LFEE191 pKa = 4.28GTRR194 pKa = 11.84FRR196 pKa = 11.84DD197 pKa = 3.21WVNPFTAKK205 pKa = 10.34LDD207 pKa = 3.75RR208 pKa = 11.84SIVTVHH214 pKa = 7.01ADD216 pKa = 3.38TVRR219 pKa = 11.84TVQSNNPNGTYY230 pKa = 10.2KK231 pKa = 10.39VYY233 pKa = 10.75KK234 pKa = 9.7RR235 pKa = 11.84YY236 pKa = 10.34YY237 pKa = 8.05PINRR241 pKa = 11.84GIVYY245 pKa = 10.49ADD247 pKa = 4.05DD248 pKa = 3.63EE249 pKa = 4.81SGAGKK254 pKa = 10.6KK255 pKa = 9.98NDD257 pKa = 4.2AIYY260 pKa = 10.51AAGTTKK266 pKa = 10.7GNSGDD271 pKa = 4.48LFVLDD276 pKa = 4.06MFSAVGDD283 pKa = 4.07NEE285 pKa = 4.4GSTLNFGVHH294 pKa = 6.13ARR296 pKa = 11.84YY297 pKa = 8.98YY298 pKa = 7.44WHH300 pKa = 7.27EE301 pKa = 4.21GSGQQ305 pKa = 3.18
Molecular weight: 34.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
611 |
305 |
306 |
305.5 |
34.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.71 ± 0.983 | 1.8 ± 0.323 |
6.056 ± 0.535 | 3.928 ± 0.429 |
4.91 ± 0.645 | 9.493 ± 0.01 |
2.619 ± 0.864 | 4.583 ± 0.429 |
3.764 ± 0.104 | 6.219 ± 0.86 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.473 ± 0.11 | 4.583 ± 0.438 |
3.928 ± 0.438 | 2.946 ± 0.647 |
9.82 ± 1.528 | 8.674 ± 0.316 |
5.565 ± 0.656 | 5.728 ± 0.982 |
2.455 ± 0.322 | 4.746 ± 0.33 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
