
Rhinolophus simulator polyomavirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; unclassified Polyomaviridae
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
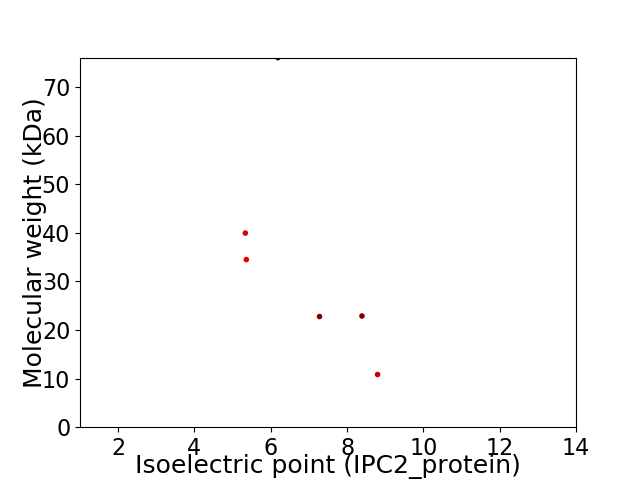
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
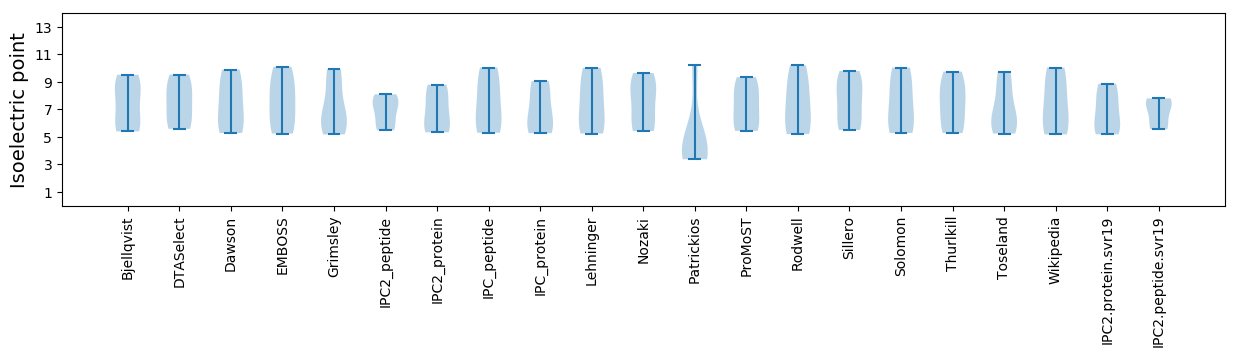
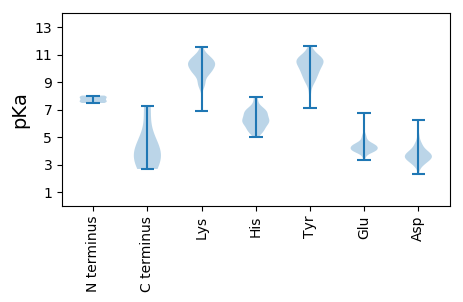
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A223YSI5|A0A223YSI5_9POLY Minor capsid protein VP2 OS=Rhinolophus simulator polyomavirus 3 OX=2029306 GN=VP3 PE=3 SV=1
MM1 pKa = 7.48APKK4 pKa = 9.99RR5 pKa = 11.84KK6 pKa = 9.71EE7 pKa = 4.15GEE9 pKa = 4.26CSKK12 pKa = 10.83SCPKK16 pKa = 10.32PSVVPRR22 pKa = 11.84LIVKK26 pKa = 10.26GGIEE30 pKa = 4.14VLSVKK35 pKa = 9.61TGPDD39 pKa = 2.85STTQIEE45 pKa = 4.84AYY47 pKa = 9.9LNPRR51 pKa = 11.84MGHH54 pKa = 5.88NLPTDD59 pKa = 3.23EE60 pKa = 5.22RR61 pKa = 11.84YY62 pKa = 10.39GYY64 pKa = 10.36SDD66 pKa = 3.56NVTVATSHH74 pKa = 6.74TDD76 pKa = 3.53DD77 pKa = 3.74NPKK80 pKa = 9.9IAEE83 pKa = 4.29LPTYY87 pKa = 9.88SAARR91 pKa = 11.84IALPMLNDD99 pKa = 4.69DD100 pKa = 4.4MTCSTLQMWEE110 pKa = 3.93AVSVKK115 pKa = 9.9TEE117 pKa = 3.91VVGASSLINGHH128 pKa = 5.5MFGKK132 pKa = 10.22RR133 pKa = 11.84VNNDD137 pKa = 2.98YY138 pKa = 11.48GIATPIEE145 pKa = 4.07GMNFHH150 pKa = 6.75MFAVGGEE157 pKa = 4.05PLEE160 pKa = 4.34LQAVVNNSRR169 pKa = 11.84TTWPAGTIGPKK180 pKa = 10.41GNSPKK185 pKa = 10.71LQVLDD190 pKa = 3.95PTAKK194 pKa = 10.61AKK196 pKa = 10.37LDD198 pKa = 3.49KK199 pKa = 11.2DD200 pKa = 3.44GAYY203 pKa = 9.58PIEE206 pKa = 4.6AWSPDD211 pKa = 3.13PSKK214 pKa = 11.56NEE216 pKa = 3.36NTRR219 pKa = 11.84YY220 pKa = 9.17YY221 pKa = 11.16GSYY224 pKa = 9.72TGGTTTPPVVQFTNTVTTVLLDD246 pKa = 3.84EE247 pKa = 5.12NGVGPLCKK255 pKa = 10.08GDD257 pKa = 4.05GLFLTAADD265 pKa = 3.01IVGFFTDD272 pKa = 2.94SSGYY276 pKa = 8.77QSYY279 pKa = 10.8RR280 pKa = 11.84GLPRR284 pKa = 11.84YY285 pKa = 10.0FNVQLRR291 pKa = 11.84KK292 pKa = 9.61RR293 pKa = 11.84VVKK296 pKa = 10.6NPYY299 pKa = 9.32PVTSLLSSLFCNLMPQITGQPMEE322 pKa = 4.75GNDD325 pKa = 3.84GQVEE329 pKa = 4.38EE330 pKa = 4.24VRR332 pKa = 11.84VYY334 pKa = 10.65QGLEE338 pKa = 4.21GVSGDD343 pKa = 3.8PDD345 pKa = 3.27MEE347 pKa = 4.37RR348 pKa = 11.84YY349 pKa = 8.96VDD351 pKa = 4.97KK352 pKa = 10.99FGQEE356 pKa = 3.75QTNIPGGACIRR367 pKa = 11.84PP368 pKa = 3.6
MM1 pKa = 7.48APKK4 pKa = 9.99RR5 pKa = 11.84KK6 pKa = 9.71EE7 pKa = 4.15GEE9 pKa = 4.26CSKK12 pKa = 10.83SCPKK16 pKa = 10.32PSVVPRR22 pKa = 11.84LIVKK26 pKa = 10.26GGIEE30 pKa = 4.14VLSVKK35 pKa = 9.61TGPDD39 pKa = 2.85STTQIEE45 pKa = 4.84AYY47 pKa = 9.9LNPRR51 pKa = 11.84MGHH54 pKa = 5.88NLPTDD59 pKa = 3.23EE60 pKa = 5.22RR61 pKa = 11.84YY62 pKa = 10.39GYY64 pKa = 10.36SDD66 pKa = 3.56NVTVATSHH74 pKa = 6.74TDD76 pKa = 3.53DD77 pKa = 3.74NPKK80 pKa = 9.9IAEE83 pKa = 4.29LPTYY87 pKa = 9.88SAARR91 pKa = 11.84IALPMLNDD99 pKa = 4.69DD100 pKa = 4.4MTCSTLQMWEE110 pKa = 3.93AVSVKK115 pKa = 9.9TEE117 pKa = 3.91VVGASSLINGHH128 pKa = 5.5MFGKK132 pKa = 10.22RR133 pKa = 11.84VNNDD137 pKa = 2.98YY138 pKa = 11.48GIATPIEE145 pKa = 4.07GMNFHH150 pKa = 6.75MFAVGGEE157 pKa = 4.05PLEE160 pKa = 4.34LQAVVNNSRR169 pKa = 11.84TTWPAGTIGPKK180 pKa = 10.41GNSPKK185 pKa = 10.71LQVLDD190 pKa = 3.95PTAKK194 pKa = 10.61AKK196 pKa = 10.37LDD198 pKa = 3.49KK199 pKa = 11.2DD200 pKa = 3.44GAYY203 pKa = 9.58PIEE206 pKa = 4.6AWSPDD211 pKa = 3.13PSKK214 pKa = 11.56NEE216 pKa = 3.36NTRR219 pKa = 11.84YY220 pKa = 9.17YY221 pKa = 11.16GSYY224 pKa = 9.72TGGTTTPPVVQFTNTVTTVLLDD246 pKa = 3.84EE247 pKa = 5.12NGVGPLCKK255 pKa = 10.08GDD257 pKa = 4.05GLFLTAADD265 pKa = 3.01IVGFFTDD272 pKa = 2.94SSGYY276 pKa = 8.77QSYY279 pKa = 10.8RR280 pKa = 11.84GLPRR284 pKa = 11.84YY285 pKa = 10.0FNVQLRR291 pKa = 11.84KK292 pKa = 9.61RR293 pKa = 11.84VVKK296 pKa = 10.6NPYY299 pKa = 9.32PVTSLLSSLFCNLMPQITGQPMEE322 pKa = 4.75GNDD325 pKa = 3.84GQVEE329 pKa = 4.38EE330 pKa = 4.24VRR332 pKa = 11.84VYY334 pKa = 10.65QGLEE338 pKa = 4.21GVSGDD343 pKa = 3.8PDD345 pKa = 3.27MEE347 pKa = 4.37RR348 pKa = 11.84YY349 pKa = 8.96VDD351 pKa = 4.97KK352 pKa = 10.99FGQEE356 pKa = 3.75QTNIPGGACIRR367 pKa = 11.84PP368 pKa = 3.6
Molecular weight: 39.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A223Z975|A0A223Z975_9POLY Small T antigen OS=Rhinolophus simulator polyomavirus 3 OX=2029306 PE=4 SV=1
MM1 pKa = 8.02DD2 pKa = 6.23LLHH5 pKa = 6.73GSSGGMTSTEE15 pKa = 3.76NGMTSSVMRR24 pKa = 11.84PSAAVMKK31 pKa = 10.34RR32 pKa = 11.84SNLEE36 pKa = 3.49EE37 pKa = 4.54DD38 pKa = 3.62PAEE41 pKa = 4.0IALNMIVGSTLDD53 pKa = 3.69PLRR56 pKa = 11.84GPLLHH61 pKa = 7.04PLNPRR66 pKa = 11.84NKK68 pKa = 9.28KK69 pKa = 8.82PSMYY73 pKa = 10.48LRR75 pKa = 11.84IFLVSLIVSLVMLFIVIRR93 pKa = 11.84QFVHH97 pKa = 6.47FF98 pKa = 4.64
MM1 pKa = 8.02DD2 pKa = 6.23LLHH5 pKa = 6.73GSSGGMTSTEE15 pKa = 3.76NGMTSSVMRR24 pKa = 11.84PSAAVMKK31 pKa = 10.34RR32 pKa = 11.84SNLEE36 pKa = 3.49EE37 pKa = 4.54DD38 pKa = 3.62PAEE41 pKa = 4.0IALNMIVGSTLDD53 pKa = 3.69PLRR56 pKa = 11.84GPLLHH61 pKa = 7.04PLNPRR66 pKa = 11.84NKK68 pKa = 9.28KK69 pKa = 8.82PSMYY73 pKa = 10.48LRR75 pKa = 11.84IFLVSLIVSLVMLFIVIRR93 pKa = 11.84QFVHH97 pKa = 6.47FF98 pKa = 4.64
Molecular weight: 10.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1827 |
98 |
663 |
304.5 |
34.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.185 ± 1.377 | 2.189 ± 0.889 |
4.926 ± 0.528 | 6.897 ± 0.411 |
3.941 ± 0.755 | 6.568 ± 0.834 |
2.518 ± 0.403 | 4.324 ± 0.225 |
6.513 ± 1.74 | 9.852 ± 0.797 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.175 ± 0.522 | 4.488 ± 0.539 |
5.802 ± 0.606 | 3.612 ± 0.389 |
4.762 ± 0.997 | 6.24 ± 0.487 |
5.473 ± 0.671 | 6.623 ± 0.642 |
1.861 ± 0.486 | 4.05 ± 0.277 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
