
Ferrovibrio terrae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Ferrovibrio
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
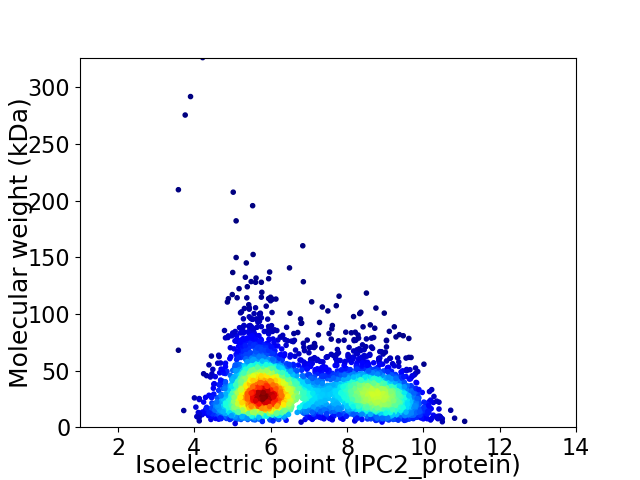
Virtual 2D-PAGE plot for 4107 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A516H517|A0A516H517_9PROT Protein TonB OS=Ferrovibrio terrae OX=2594003 GN=FNB15_16760 PE=3 SV=1
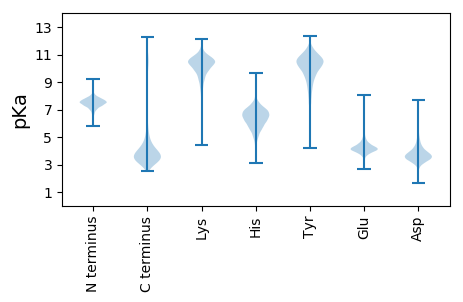
MM1 pKa = 7.57SPATDD6 pKa = 3.43SDD8 pKa = 3.93SGGEE12 pKa = 3.97KK13 pKa = 9.5MAVITGDD20 pKa = 3.1ATDD23 pKa = 3.57NVLNGTAAADD33 pKa = 3.5QIYY36 pKa = 10.08GLNGNDD42 pKa = 3.57TLTGGNGIDD51 pKa = 4.76LLDD54 pKa = 4.84GGDD57 pKa = 4.61GDD59 pKa = 4.71DD60 pKa = 4.47SLYY63 pKa = 11.29GGNQNDD69 pKa = 4.21TLNGGNGIDD78 pKa = 3.2TAVFEE83 pKa = 4.66KK84 pKa = 10.72PIGRR88 pKa = 11.84YY89 pKa = 9.55RR90 pKa = 11.84ITYY93 pKa = 8.01NNGVITVDD101 pKa = 3.27AQSGYY106 pKa = 11.03EE107 pKa = 4.44DD108 pKa = 4.2VDD110 pKa = 3.74TLTGMEE116 pKa = 4.01KK117 pKa = 10.66LKK119 pKa = 11.13FNGLVYY125 pKa = 10.76DD126 pKa = 3.92FTGWTGGPLPEE137 pKa = 4.23PTMPAGTVYY146 pKa = 9.25GTEE149 pKa = 3.85GHH151 pKa = 5.77EE152 pKa = 4.35TLTGTGGVDD161 pKa = 3.4YY162 pKa = 11.13LHH164 pKa = 7.05GLGGDD169 pKa = 3.58DD170 pKa = 4.66VMIGGAGGDD179 pKa = 3.49ILVGGSGNDD188 pKa = 3.3TASYY192 pKa = 9.85AGSSAGVTVSLEE204 pKa = 4.2TGLGFYY210 pKa = 10.91GDD212 pKa = 3.79AANDD216 pKa = 3.51VLHH219 pKa = 6.77GFEE222 pKa = 4.44NLQGSSFNDD231 pKa = 3.33TLTGNGGDD239 pKa = 4.12NIFISSGGVDD249 pKa = 5.26DD250 pKa = 3.82ITGGAGVDD258 pKa = 3.05WVDD261 pKa = 3.34YY262 pKa = 11.3SDD264 pKa = 3.36IVKK267 pKa = 9.37PAGMTGMFFQRR278 pKa = 11.84GTPTNIASYY287 pKa = 9.81FYY289 pKa = 10.27NASINPFKK297 pKa = 10.53DD298 pKa = 2.77TLRR301 pKa = 11.84QVEE304 pKa = 4.42GVRR307 pKa = 11.84GSAYY311 pKa = 10.55DD312 pKa = 3.67DD313 pKa = 3.66VYY315 pKa = 11.5MNILTSDD322 pKa = 3.57DD323 pKa = 3.83MFEE326 pKa = 4.74GGAGADD332 pKa = 3.19IFDD335 pKa = 3.97GRR337 pKa = 11.84GGNDD341 pKa = 3.08TLVYY345 pKa = 8.12THH347 pKa = 6.82SAAGVNVNLATGAASGGDD365 pKa = 3.28ATGDD369 pKa = 3.53QFTGSYY375 pKa = 7.2VWRR378 pKa = 11.84GTTRR382 pKa = 11.84NLAGATNLSGSEE394 pKa = 4.12HH395 pKa = 7.42ADD397 pKa = 3.24TLTGSSVANVLRR409 pKa = 11.84GADD412 pKa = 3.48GDD414 pKa = 3.89DD415 pKa = 3.53TLRR418 pKa = 11.84GDD420 pKa = 4.51GGNDD424 pKa = 3.39TLDD427 pKa = 3.52GGAGVDD433 pKa = 3.14IAVYY437 pKa = 10.46ALSLNNYY444 pKa = 6.86TISYY448 pKa = 9.23NSGAITVAANIGTEE462 pKa = 4.28GTDD465 pKa = 3.0TLTGIEE471 pKa = 4.63KK472 pKa = 10.82LLFANGTLDD481 pKa = 3.45LTEE484 pKa = 4.17WTGGPLPPLLGPNDD498 pKa = 3.4IMGSSGDD505 pKa = 3.65NVLTGTAAGQSIYY518 pKa = 11.23GMAGNDD524 pKa = 3.99SISGAGGNDD533 pKa = 3.22VLTGGDD539 pKa = 3.52GSDD542 pKa = 3.35TYY544 pKa = 11.54LVARR548 pKa = 11.84GDD550 pKa = 3.91GVDD553 pKa = 5.37DD554 pKa = 3.96IVQAGITDD562 pKa = 3.74AAGTTDD568 pKa = 3.5AVLFGSGVSYY578 pKa = 10.67DD579 pKa = 3.77QLWFRR584 pKa = 11.84QVGNDD589 pKa = 3.93LRR591 pKa = 11.84IDD593 pKa = 4.04VIGEE597 pKa = 4.15SASSVLLKK605 pKa = 10.3SWYY608 pKa = 8.16TDD610 pKa = 2.8STRR613 pKa = 11.84RR614 pKa = 11.84VDD616 pKa = 5.15SIQTTDD622 pKa = 3.26GSHH625 pKa = 6.25SLSAANVQALVDD637 pKa = 3.91AMATYY642 pKa = 7.6TQPGTGDD649 pKa = 3.5FTLSPQLASDD659 pKa = 4.6LAPVFATTWAA669 pKa = 3.56
MM1 pKa = 7.57SPATDD6 pKa = 3.43SDD8 pKa = 3.93SGGEE12 pKa = 3.97KK13 pKa = 9.5MAVITGDD20 pKa = 3.1ATDD23 pKa = 3.57NVLNGTAAADD33 pKa = 3.5QIYY36 pKa = 10.08GLNGNDD42 pKa = 3.57TLTGGNGIDD51 pKa = 4.76LLDD54 pKa = 4.84GGDD57 pKa = 4.61GDD59 pKa = 4.71DD60 pKa = 4.47SLYY63 pKa = 11.29GGNQNDD69 pKa = 4.21TLNGGNGIDD78 pKa = 3.2TAVFEE83 pKa = 4.66KK84 pKa = 10.72PIGRR88 pKa = 11.84YY89 pKa = 9.55RR90 pKa = 11.84ITYY93 pKa = 8.01NNGVITVDD101 pKa = 3.27AQSGYY106 pKa = 11.03EE107 pKa = 4.44DD108 pKa = 4.2VDD110 pKa = 3.74TLTGMEE116 pKa = 4.01KK117 pKa = 10.66LKK119 pKa = 11.13FNGLVYY125 pKa = 10.76DD126 pKa = 3.92FTGWTGGPLPEE137 pKa = 4.23PTMPAGTVYY146 pKa = 9.25GTEE149 pKa = 3.85GHH151 pKa = 5.77EE152 pKa = 4.35TLTGTGGVDD161 pKa = 3.4YY162 pKa = 11.13LHH164 pKa = 7.05GLGGDD169 pKa = 3.58DD170 pKa = 4.66VMIGGAGGDD179 pKa = 3.49ILVGGSGNDD188 pKa = 3.3TASYY192 pKa = 9.85AGSSAGVTVSLEE204 pKa = 4.2TGLGFYY210 pKa = 10.91GDD212 pKa = 3.79AANDD216 pKa = 3.51VLHH219 pKa = 6.77GFEE222 pKa = 4.44NLQGSSFNDD231 pKa = 3.33TLTGNGGDD239 pKa = 4.12NIFISSGGVDD249 pKa = 5.26DD250 pKa = 3.82ITGGAGVDD258 pKa = 3.05WVDD261 pKa = 3.34YY262 pKa = 11.3SDD264 pKa = 3.36IVKK267 pKa = 9.37PAGMTGMFFQRR278 pKa = 11.84GTPTNIASYY287 pKa = 9.81FYY289 pKa = 10.27NASINPFKK297 pKa = 10.53DD298 pKa = 2.77TLRR301 pKa = 11.84QVEE304 pKa = 4.42GVRR307 pKa = 11.84GSAYY311 pKa = 10.55DD312 pKa = 3.67DD313 pKa = 3.66VYY315 pKa = 11.5MNILTSDD322 pKa = 3.57DD323 pKa = 3.83MFEE326 pKa = 4.74GGAGADD332 pKa = 3.19IFDD335 pKa = 3.97GRR337 pKa = 11.84GGNDD341 pKa = 3.08TLVYY345 pKa = 8.12THH347 pKa = 6.82SAAGVNVNLATGAASGGDD365 pKa = 3.28ATGDD369 pKa = 3.53QFTGSYY375 pKa = 7.2VWRR378 pKa = 11.84GTTRR382 pKa = 11.84NLAGATNLSGSEE394 pKa = 4.12HH395 pKa = 7.42ADD397 pKa = 3.24TLTGSSVANVLRR409 pKa = 11.84GADD412 pKa = 3.48GDD414 pKa = 3.89DD415 pKa = 3.53TLRR418 pKa = 11.84GDD420 pKa = 4.51GGNDD424 pKa = 3.39TLDD427 pKa = 3.52GGAGVDD433 pKa = 3.14IAVYY437 pKa = 10.46ALSLNNYY444 pKa = 6.86TISYY448 pKa = 9.23NSGAITVAANIGTEE462 pKa = 4.28GTDD465 pKa = 3.0TLTGIEE471 pKa = 4.63KK472 pKa = 10.82LLFANGTLDD481 pKa = 3.45LTEE484 pKa = 4.17WTGGPLPPLLGPNDD498 pKa = 3.4IMGSSGDD505 pKa = 3.65NVLTGTAAGQSIYY518 pKa = 11.23GMAGNDD524 pKa = 3.99SISGAGGNDD533 pKa = 3.22VLTGGDD539 pKa = 3.52GSDD542 pKa = 3.35TYY544 pKa = 11.54LVARR548 pKa = 11.84GDD550 pKa = 3.91GVDD553 pKa = 5.37DD554 pKa = 3.96IVQAGITDD562 pKa = 3.74AAGTTDD568 pKa = 3.5AVLFGSGVSYY578 pKa = 10.67DD579 pKa = 3.77QLWFRR584 pKa = 11.84QVGNDD589 pKa = 3.93LRR591 pKa = 11.84IDD593 pKa = 4.04VIGEE597 pKa = 4.15SASSVLLKK605 pKa = 10.3SWYY608 pKa = 8.16TDD610 pKa = 2.8STRR613 pKa = 11.84RR614 pKa = 11.84VDD616 pKa = 5.15SIQTTDD622 pKa = 3.26GSHH625 pKa = 6.25SLSAANVQALVDD637 pKa = 3.91AMATYY642 pKa = 7.6TQPGTGDD649 pKa = 3.5FTLSPQLASDD659 pKa = 4.6LAPVFATTWAA669 pKa = 3.56
Molecular weight: 67.96 kDa
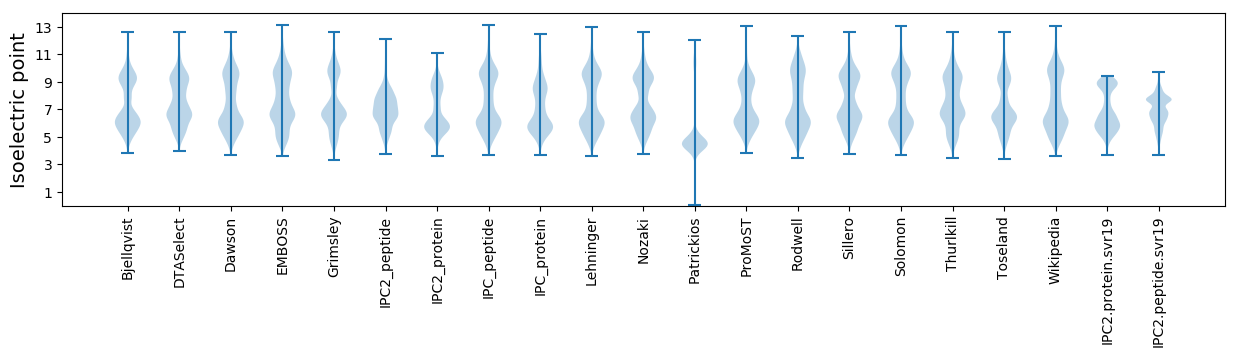
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A516H194|A0A516H194_9PROT 3-alpha 7-alpha 12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase OS=Ferrovibrio terrae OX=2594003 GN=FNB15_09755 PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.27QPSKK10 pKa = 9.73LVRR13 pKa = 11.84KK14 pKa = 9.15RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.42GFRR20 pKa = 11.84ARR22 pKa = 11.84MATVGGRR29 pKa = 11.84NVIASRR35 pKa = 11.84RR36 pKa = 11.84AQGRR40 pKa = 11.84KK41 pKa = 9.31KK42 pKa = 10.65LSAA45 pKa = 3.91
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.27QPSKK10 pKa = 9.73LVRR13 pKa = 11.84KK14 pKa = 9.15RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.42GFRR20 pKa = 11.84ARR22 pKa = 11.84MATVGGRR29 pKa = 11.84NVIASRR35 pKa = 11.84RR36 pKa = 11.84AQGRR40 pKa = 11.84KK41 pKa = 9.31KK42 pKa = 10.65LSAA45 pKa = 3.91
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1303169 |
29 |
3226 |
317.3 |
34.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.612 ± 0.055 | 0.84 ± 0.011 |
5.528 ± 0.028 | 5.232 ± 0.034 |
3.649 ± 0.024 | 8.481 ± 0.039 |
2.044 ± 0.021 | 5.168 ± 0.028 |
3.698 ± 0.035 | 10.576 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.628 ± 0.02 | 2.697 ± 0.02 |
5.192 ± 0.038 | 3.614 ± 0.032 |
6.844 ± 0.038 | 5.073 ± 0.027 |
5.284 ± 0.054 | 7.229 ± 0.032 |
1.339 ± 0.015 | 2.271 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
