
Candidatus Berkiella aquae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Legionellales; Coxiellaceae; Candidatus Berkiella
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
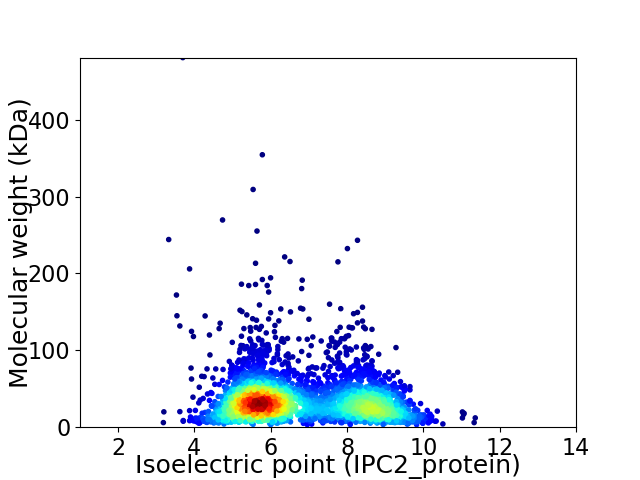
Virtual 2D-PAGE plot for 3170 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q9Z102|A0A0Q9Z102_9COXI Uncharacterized protein OS=Candidatus Berkiella aquae OX=295108 GN=HT99x_00261 PE=4 SV=1
MM1 pKa = 7.39EE2 pKa = 5.86RR3 pKa = 11.84NPEE6 pKa = 4.37RR7 pKa = 11.84IEE9 pKa = 4.11HH10 pKa = 6.24DD11 pKa = 2.87HH12 pKa = 7.42DD13 pKa = 3.64IFVPTVEE20 pKa = 5.01AVFPYY25 pKa = 10.42EE26 pKa = 3.83KK27 pKa = 10.42LAVPFEE33 pKa = 4.13KK34 pKa = 10.67SIDD37 pKa = 3.72SYY39 pKa = 11.79KK40 pKa = 11.03VLDD43 pKa = 3.92EE44 pKa = 4.18EE45 pKa = 5.44ALLKK49 pKa = 10.53VANQTSKK56 pKa = 10.87QSDD59 pKa = 4.01EE60 pKa = 4.42EE61 pKa = 4.82DD62 pKa = 3.43VFAPSLEE69 pKa = 4.05GVFPYY74 pKa = 10.48EE75 pKa = 3.89VLASSFANAMDD86 pKa = 3.95KK87 pKa = 11.17YY88 pKa = 11.08VVLDD92 pKa = 3.88EE93 pKa = 4.54AALLKK98 pKa = 10.14TIEE101 pKa = 4.32DD102 pKa = 3.52AVAKK106 pKa = 10.54SKK108 pKa = 11.28LNANEE113 pKa = 4.31NEE115 pKa = 3.92FLDD118 pKa = 4.67VKK120 pKa = 11.12FEE122 pKa = 4.91DD123 pKa = 4.14LFQIPVTEE131 pKa = 4.18GNAYY135 pKa = 9.03NIKK138 pKa = 8.79MAQYY142 pKa = 8.12MKK144 pKa = 10.07EE145 pKa = 3.84GRR147 pKa = 11.84VEE149 pKa = 4.08SSGSQLEE156 pKa = 4.48VVSVNFDD163 pKa = 3.47YY164 pKa = 11.3QPIEE168 pKa = 3.93MLDD171 pKa = 3.68LVISVPAPQPSITTTTTTNVQSPPTVTTNPQHH203 pKa = 6.56GLPTAPAITMPDD215 pKa = 3.1IFEE218 pKa = 4.29VTLDD222 pKa = 3.58TVAFTTSIGNIFTKK236 pKa = 10.47GASFGPSGGNLVNVYY251 pKa = 9.83FGLPIRR257 pKa = 11.84PGSSEE262 pKa = 3.45IRR264 pKa = 11.84NSDD267 pKa = 3.56VVTLTTAEE275 pKa = 4.06GNEE278 pKa = 3.96LNFYY282 pKa = 10.83LSTFNGHH289 pKa = 6.13QIGDD293 pKa = 3.59YY294 pKa = 10.7EE295 pKa = 4.94YY296 pKa = 9.45ILHH299 pKa = 6.56HH300 pKa = 6.72AVPHH304 pKa = 5.94LLGNPLYY311 pKa = 10.7DD312 pKa = 4.18VIQMGGTKK320 pKa = 9.59IFFDD324 pKa = 3.53NFVYY328 pKa = 10.7SLSNTYY334 pKa = 10.86LEE336 pKa = 4.59TNIGTLTFPIIDD348 pKa = 4.52DD349 pKa = 3.92VPLATSQNGGTISEE363 pKa = 4.17AAIEE367 pKa = 4.16NSGTDD372 pKa = 3.51QSHH375 pKa = 5.52VPALLSGTLINPPVDD390 pKa = 3.02RR391 pKa = 11.84FGADD395 pKa = 2.84GGTVTSVTINGGSYY409 pKa = 10.87GFLFGHH415 pKa = 6.35IVVITTEE422 pKa = 4.35GNILTVNVNTGAYY435 pKa = 7.88TFLLSNPLHH444 pKa = 5.76NTNNQPINQVFTYY457 pKa = 10.68VFTDD461 pKa = 3.09SDD463 pKa = 3.95GSTASNTLTITIDD476 pKa = 3.54DD477 pKa = 5.74DD478 pKa = 4.15IPIATEE484 pKa = 3.81KK485 pKa = 10.51TNTASEE491 pKa = 4.37TLCFVNGSDD500 pKa = 3.36TATGNLISDD509 pKa = 4.2DD510 pKa = 3.91NGFGISLFGADD521 pKa = 4.52GGDD524 pKa = 2.9ITAVNGVTDD533 pKa = 4.01ASDD536 pKa = 3.62GLVDD540 pKa = 5.35GIIHH544 pKa = 7.22APTTFGDD551 pKa = 3.42IEE553 pKa = 4.88VYY555 pKa = 10.41VAVQSGHH562 pKa = 5.95QIGDD566 pKa = 3.61YY567 pKa = 10.24IYY569 pKa = 10.53TLDD572 pKa = 3.63TSKK575 pKa = 9.51TAPQNDD581 pKa = 3.54NLITVLDD588 pKa = 4.54TISYY592 pKa = 8.1TITDD596 pKa = 3.49NDD598 pKa = 3.8GSQDD602 pKa = 3.51SANLVVTVTLNQAPTAVDD620 pKa = 3.78DD621 pKa = 5.0VGTTDD626 pKa = 3.85EE627 pKa = 4.16NTILDD632 pKa = 4.09VLTVNGVLSNDD643 pKa = 3.44TDD645 pKa = 4.32PNPGDD650 pKa = 3.68TKK652 pKa = 10.61IVSAVNGLGTNVGNQFTLLSNALLLLNADD681 pKa = 3.58GSYY684 pKa = 10.03TYY686 pKa = 11.04NPNGVFNYY694 pKa = 9.8LAAGSQGIDD703 pKa = 2.79SFTYY707 pKa = 9.75TMHH710 pKa = 7.28DD711 pKa = 3.45AEE713 pKa = 5.06GLSSSATVTITINGVNSAPVAVDD736 pKa = 4.0DD737 pKa = 5.91SNTTDD742 pKa = 3.25ANAIINVNTIVDD754 pKa = 3.73PHH756 pKa = 6.93NLLINDD762 pKa = 3.93TDD764 pKa = 4.15DD765 pKa = 3.99VGDD768 pKa = 3.51THH770 pKa = 7.64SISEE774 pKa = 4.44VNGVAANVGNQITLASGALLTVNADD799 pKa = 2.79GTYY802 pKa = 10.53NYY804 pKa = 10.13NPNHH808 pKa = 6.1MFDD811 pKa = 4.29SLAQGSSTTDD821 pKa = 2.61SFTYY825 pKa = 9.93TLQDD829 pKa = 3.13SGGLTSTATVIITINGVNDD848 pKa = 3.38APTAVDD854 pKa = 4.96DD855 pKa = 5.7SNTTDD860 pKa = 3.71ANTMIDD866 pKa = 3.69VNLVSDD872 pKa = 3.97PHH874 pKa = 7.33NLLINDD880 pKa = 4.08TDD882 pKa = 4.38PDD884 pKa = 3.78VGDD887 pKa = 3.55TLVISEE893 pKa = 4.41VQGLAANVGVQITLASGALLTVNANGTYY921 pKa = 10.02QYY923 pKa = 10.47NPNGAFVSLSVGASTIDD940 pKa = 3.22SFTYY944 pKa = 9.54TVQDD948 pKa = 3.29SGGLTSTATVSITINGVNEE967 pKa = 3.72APIAVDD973 pKa = 4.93DD974 pKa = 4.52SNTTHH979 pKa = 6.23GTTVIDD985 pKa = 3.92VNSTLDD991 pKa = 3.73PQCLLFNDD999 pKa = 4.45SDD1001 pKa = 4.72PDD1003 pKa = 3.68TGDD1006 pKa = 3.06TFVISEE1012 pKa = 4.52VEE1014 pKa = 3.98GQAANVGVQITLTSGALLTVNADD1037 pKa = 2.8GTYY1040 pKa = 10.1QYY1042 pKa = 11.43DD1043 pKa = 3.81SNGVFAATDD1052 pKa = 3.37IDD1054 pKa = 4.11SFTYY1058 pKa = 10.41KK1059 pKa = 9.24ITDD1062 pKa = 3.37NHH1064 pKa = 6.27GLTSNAATVTINVIVPPIILDD1085 pKa = 4.01LNDD1088 pKa = 4.71DD1089 pKa = 5.24GINLIPPDD1097 pKa = 3.67EE1098 pKa = 4.22SQIAFSLFGRR1108 pKa = 11.84EE1109 pKa = 3.74NTTTIGWVNGQDD1121 pKa = 3.24GLLAIDD1127 pKa = 4.6LNGDD1131 pKa = 3.65KK1132 pKa = 10.32IINGLEE1138 pKa = 4.05EE1139 pKa = 4.16FTFTHH1144 pKa = 6.85PNAKK1148 pKa = 8.87TDD1150 pKa = 3.98LEE1152 pKa = 4.16ALRR1155 pKa = 11.84LLYY1158 pKa = 10.52DD1159 pKa = 3.82SNVDD1163 pKa = 4.77GILDD1167 pKa = 3.68MQDD1170 pKa = 3.08EE1171 pKa = 4.33DD1172 pKa = 3.48WARR1175 pKa = 11.84FGVWQDD1181 pKa = 2.99ANEE1184 pKa = 4.08NGICEE1189 pKa = 4.23PGEE1192 pKa = 3.89FLSLAEE1198 pKa = 4.56RR1199 pKa = 11.84GIASIDD1205 pKa = 4.23LISDD1209 pKa = 3.43QQSDD1213 pKa = 4.25VIAGNIIFGYY1223 pKa = 8.9ATYY1226 pKa = 8.71QTTDD1230 pKa = 3.11GQLHH1234 pKa = 5.2QLADD1238 pKa = 3.47VGLGLFF1244 pKa = 4.27
MM1 pKa = 7.39EE2 pKa = 5.86RR3 pKa = 11.84NPEE6 pKa = 4.37RR7 pKa = 11.84IEE9 pKa = 4.11HH10 pKa = 6.24DD11 pKa = 2.87HH12 pKa = 7.42DD13 pKa = 3.64IFVPTVEE20 pKa = 5.01AVFPYY25 pKa = 10.42EE26 pKa = 3.83KK27 pKa = 10.42LAVPFEE33 pKa = 4.13KK34 pKa = 10.67SIDD37 pKa = 3.72SYY39 pKa = 11.79KK40 pKa = 11.03VLDD43 pKa = 3.92EE44 pKa = 4.18EE45 pKa = 5.44ALLKK49 pKa = 10.53VANQTSKK56 pKa = 10.87QSDD59 pKa = 4.01EE60 pKa = 4.42EE61 pKa = 4.82DD62 pKa = 3.43VFAPSLEE69 pKa = 4.05GVFPYY74 pKa = 10.48EE75 pKa = 3.89VLASSFANAMDD86 pKa = 3.95KK87 pKa = 11.17YY88 pKa = 11.08VVLDD92 pKa = 3.88EE93 pKa = 4.54AALLKK98 pKa = 10.14TIEE101 pKa = 4.32DD102 pKa = 3.52AVAKK106 pKa = 10.54SKK108 pKa = 11.28LNANEE113 pKa = 4.31NEE115 pKa = 3.92FLDD118 pKa = 4.67VKK120 pKa = 11.12FEE122 pKa = 4.91DD123 pKa = 4.14LFQIPVTEE131 pKa = 4.18GNAYY135 pKa = 9.03NIKK138 pKa = 8.79MAQYY142 pKa = 8.12MKK144 pKa = 10.07EE145 pKa = 3.84GRR147 pKa = 11.84VEE149 pKa = 4.08SSGSQLEE156 pKa = 4.48VVSVNFDD163 pKa = 3.47YY164 pKa = 11.3QPIEE168 pKa = 3.93MLDD171 pKa = 3.68LVISVPAPQPSITTTTTTNVQSPPTVTTNPQHH203 pKa = 6.56GLPTAPAITMPDD215 pKa = 3.1IFEE218 pKa = 4.29VTLDD222 pKa = 3.58TVAFTTSIGNIFTKK236 pKa = 10.47GASFGPSGGNLVNVYY251 pKa = 9.83FGLPIRR257 pKa = 11.84PGSSEE262 pKa = 3.45IRR264 pKa = 11.84NSDD267 pKa = 3.56VVTLTTAEE275 pKa = 4.06GNEE278 pKa = 3.96LNFYY282 pKa = 10.83LSTFNGHH289 pKa = 6.13QIGDD293 pKa = 3.59YY294 pKa = 10.7EE295 pKa = 4.94YY296 pKa = 9.45ILHH299 pKa = 6.56HH300 pKa = 6.72AVPHH304 pKa = 5.94LLGNPLYY311 pKa = 10.7DD312 pKa = 4.18VIQMGGTKK320 pKa = 9.59IFFDD324 pKa = 3.53NFVYY328 pKa = 10.7SLSNTYY334 pKa = 10.86LEE336 pKa = 4.59TNIGTLTFPIIDD348 pKa = 4.52DD349 pKa = 3.92VPLATSQNGGTISEE363 pKa = 4.17AAIEE367 pKa = 4.16NSGTDD372 pKa = 3.51QSHH375 pKa = 5.52VPALLSGTLINPPVDD390 pKa = 3.02RR391 pKa = 11.84FGADD395 pKa = 2.84GGTVTSVTINGGSYY409 pKa = 10.87GFLFGHH415 pKa = 6.35IVVITTEE422 pKa = 4.35GNILTVNVNTGAYY435 pKa = 7.88TFLLSNPLHH444 pKa = 5.76NTNNQPINQVFTYY457 pKa = 10.68VFTDD461 pKa = 3.09SDD463 pKa = 3.95GSTASNTLTITIDD476 pKa = 3.54DD477 pKa = 5.74DD478 pKa = 4.15IPIATEE484 pKa = 3.81KK485 pKa = 10.51TNTASEE491 pKa = 4.37TLCFVNGSDD500 pKa = 3.36TATGNLISDD509 pKa = 4.2DD510 pKa = 3.91NGFGISLFGADD521 pKa = 4.52GGDD524 pKa = 2.9ITAVNGVTDD533 pKa = 4.01ASDD536 pKa = 3.62GLVDD540 pKa = 5.35GIIHH544 pKa = 7.22APTTFGDD551 pKa = 3.42IEE553 pKa = 4.88VYY555 pKa = 10.41VAVQSGHH562 pKa = 5.95QIGDD566 pKa = 3.61YY567 pKa = 10.24IYY569 pKa = 10.53TLDD572 pKa = 3.63TSKK575 pKa = 9.51TAPQNDD581 pKa = 3.54NLITVLDD588 pKa = 4.54TISYY592 pKa = 8.1TITDD596 pKa = 3.49NDD598 pKa = 3.8GSQDD602 pKa = 3.51SANLVVTVTLNQAPTAVDD620 pKa = 3.78DD621 pKa = 5.0VGTTDD626 pKa = 3.85EE627 pKa = 4.16NTILDD632 pKa = 4.09VLTVNGVLSNDD643 pKa = 3.44TDD645 pKa = 4.32PNPGDD650 pKa = 3.68TKK652 pKa = 10.61IVSAVNGLGTNVGNQFTLLSNALLLLNADD681 pKa = 3.58GSYY684 pKa = 10.03TYY686 pKa = 11.04NPNGVFNYY694 pKa = 9.8LAAGSQGIDD703 pKa = 2.79SFTYY707 pKa = 9.75TMHH710 pKa = 7.28DD711 pKa = 3.45AEE713 pKa = 5.06GLSSSATVTITINGVNSAPVAVDD736 pKa = 4.0DD737 pKa = 5.91SNTTDD742 pKa = 3.25ANAIINVNTIVDD754 pKa = 3.73PHH756 pKa = 6.93NLLINDD762 pKa = 3.93TDD764 pKa = 4.15DD765 pKa = 3.99VGDD768 pKa = 3.51THH770 pKa = 7.64SISEE774 pKa = 4.44VNGVAANVGNQITLASGALLTVNADD799 pKa = 2.79GTYY802 pKa = 10.53NYY804 pKa = 10.13NPNHH808 pKa = 6.1MFDD811 pKa = 4.29SLAQGSSTTDD821 pKa = 2.61SFTYY825 pKa = 9.93TLQDD829 pKa = 3.13SGGLTSTATVIITINGVNDD848 pKa = 3.38APTAVDD854 pKa = 4.96DD855 pKa = 5.7SNTTDD860 pKa = 3.71ANTMIDD866 pKa = 3.69VNLVSDD872 pKa = 3.97PHH874 pKa = 7.33NLLINDD880 pKa = 4.08TDD882 pKa = 4.38PDD884 pKa = 3.78VGDD887 pKa = 3.55TLVISEE893 pKa = 4.41VQGLAANVGVQITLASGALLTVNANGTYY921 pKa = 10.02QYY923 pKa = 10.47NPNGAFVSLSVGASTIDD940 pKa = 3.22SFTYY944 pKa = 9.54TVQDD948 pKa = 3.29SGGLTSTATVSITINGVNEE967 pKa = 3.72APIAVDD973 pKa = 4.93DD974 pKa = 4.52SNTTHH979 pKa = 6.23GTTVIDD985 pKa = 3.92VNSTLDD991 pKa = 3.73PQCLLFNDD999 pKa = 4.45SDD1001 pKa = 4.72PDD1003 pKa = 3.68TGDD1006 pKa = 3.06TFVISEE1012 pKa = 4.52VEE1014 pKa = 3.98GQAANVGVQITLTSGALLTVNADD1037 pKa = 2.8GTYY1040 pKa = 10.1QYY1042 pKa = 11.43DD1043 pKa = 3.81SNGVFAATDD1052 pKa = 3.37IDD1054 pKa = 4.11SFTYY1058 pKa = 10.41KK1059 pKa = 9.24ITDD1062 pKa = 3.37NHH1064 pKa = 6.27GLTSNAATVTINVIVPPIILDD1085 pKa = 4.01LNDD1088 pKa = 4.71DD1089 pKa = 5.24GINLIPPDD1097 pKa = 3.67EE1098 pKa = 4.22SQIAFSLFGRR1108 pKa = 11.84EE1109 pKa = 3.74NTTTIGWVNGQDD1121 pKa = 3.24GLLAIDD1127 pKa = 4.6LNGDD1131 pKa = 3.65KK1132 pKa = 10.32IINGLEE1138 pKa = 4.05EE1139 pKa = 4.16FTFTHH1144 pKa = 6.85PNAKK1148 pKa = 8.87TDD1150 pKa = 3.98LEE1152 pKa = 4.16ALRR1155 pKa = 11.84LLYY1158 pKa = 10.52DD1159 pKa = 3.82SNVDD1163 pKa = 4.77GILDD1167 pKa = 3.68MQDD1170 pKa = 3.08EE1171 pKa = 4.33DD1172 pKa = 3.48WARR1175 pKa = 11.84FGVWQDD1181 pKa = 2.99ANEE1184 pKa = 4.08NGICEE1189 pKa = 4.23PGEE1192 pKa = 3.89FLSLAEE1198 pKa = 4.56RR1199 pKa = 11.84GIASIDD1205 pKa = 4.23LISDD1209 pKa = 3.43QQSDD1213 pKa = 4.25VIAGNIIFGYY1223 pKa = 8.9ATYY1226 pKa = 8.71QTTDD1230 pKa = 3.11GQLHH1234 pKa = 5.2QLADD1238 pKa = 3.47VGLGLFF1244 pKa = 4.27
Molecular weight: 131.73 kDa
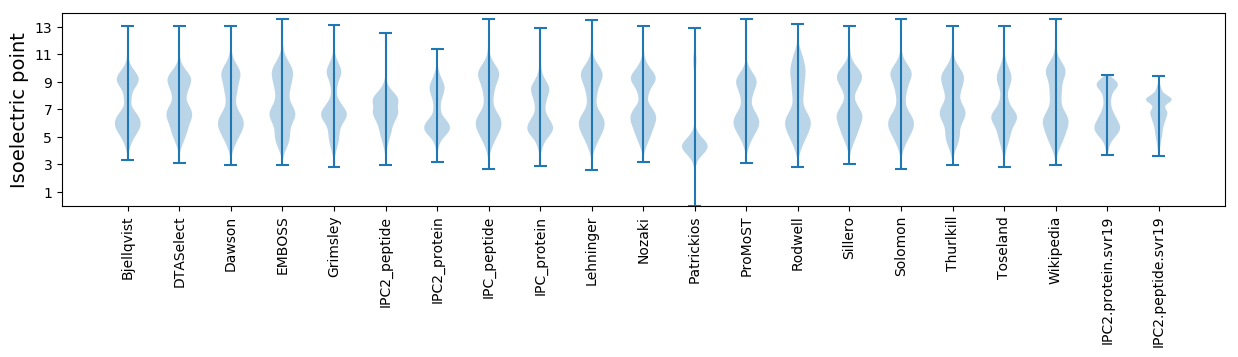
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q9YZE6|A0A0Q9YZE6_9COXI Methylmalonyl-CoA carboxyltransferase 12S subunit OS=Candidatus Berkiella aquae OX=295108 GN=HT99x_00826 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 10.36RR3 pKa = 11.84PFQRR7 pKa = 11.84KK8 pKa = 7.04SQVRR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84THH17 pKa = 6.09GFRR20 pKa = 11.84QRR22 pKa = 11.84MQTKK26 pKa = 9.63GGRR29 pKa = 11.84AVLANRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.99GRR40 pKa = 11.84KK41 pKa = 8.45RR42 pKa = 11.84LAPVGGTKK50 pKa = 10.05KK51 pKa = 10.67
MM1 pKa = 7.44KK2 pKa = 10.36RR3 pKa = 11.84PFQRR7 pKa = 11.84KK8 pKa = 7.04SQVRR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84THH17 pKa = 6.09GFRR20 pKa = 11.84QRR22 pKa = 11.84MQTKK26 pKa = 9.63GGRR29 pKa = 11.84AVLANRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.99GRR40 pKa = 11.84KK41 pKa = 8.45RR42 pKa = 11.84LAPVGGTKK50 pKa = 10.05KK51 pKa = 10.67
Molecular weight: 6.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1075553 |
29 |
4642 |
339.3 |
38.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.282 ± 0.046 | 1.096 ± 0.019 |
4.886 ± 0.04 | 6.174 ± 0.053 |
4.212 ± 0.035 | 5.889 ± 0.052 |
2.563 ± 0.024 | 7.135 ± 0.042 |
6.527 ± 0.063 | 10.725 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.44 ± 0.02 | 4.763 ± 0.038 |
4.253 ± 0.033 | 4.995 ± 0.043 |
4.202 ± 0.037 | 6.302 ± 0.037 |
5.41 ± 0.042 | 5.898 ± 0.039 |
1.078 ± 0.016 | 3.169 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
