
Brown greater galago prosimian foamy virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Spumaretrovirinae; Prosimiispumavirus
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
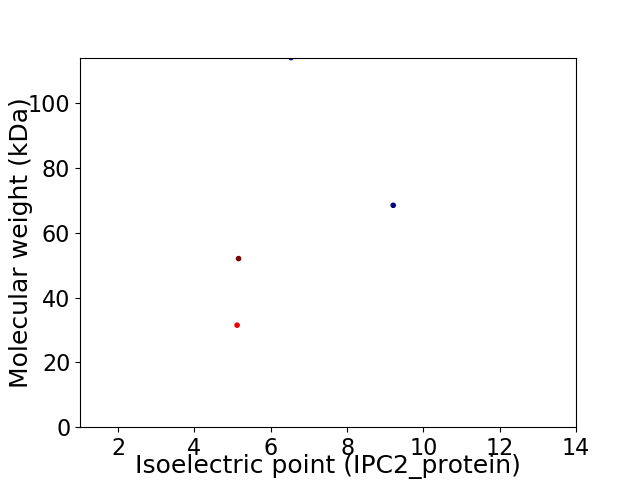
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
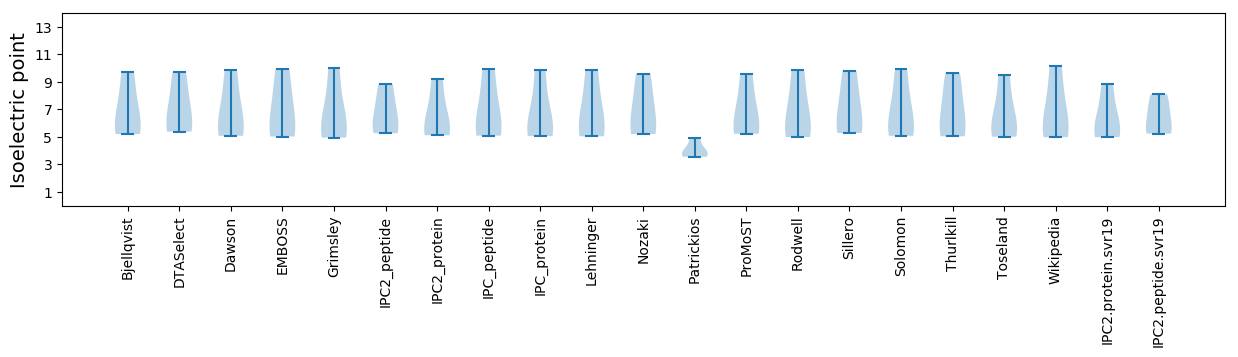
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A088FA16|A0A088FA16_9RETR Bel1 protein OS=Brown greater galago prosimian foamy virus OX=2170139 GN=bel1 PE=4 SV=1
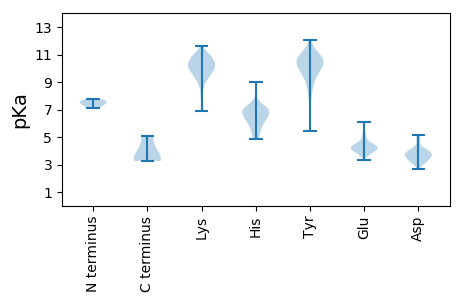
MM1 pKa = 7.52ASWNPGQKK9 pKa = 10.27GINIPQNDD17 pKa = 4.32EE18 pKa = 3.87SDD20 pKa = 3.86LPQPRR25 pKa = 11.84PLPALPASEE34 pKa = 5.13FMDD37 pKa = 3.58HH38 pKa = 6.77NYY40 pKa = 10.52VGMRR44 pKa = 11.84QWYY47 pKa = 8.32NDD49 pKa = 3.56PSGGYY54 pKa = 8.49PDD56 pKa = 4.9LGNEE60 pKa = 4.12EE61 pKa = 4.16LHH63 pKa = 7.43DD64 pKa = 3.98VLFSLAYY71 pKa = 10.16NCLHH75 pKa = 7.16FYY77 pKa = 10.7KK78 pKa = 10.26SLPIVRR84 pKa = 11.84QRR86 pKa = 11.84FKK88 pKa = 11.1CYY90 pKa = 10.32LEE92 pKa = 4.56PSFQRR97 pKa = 11.84VGIYY101 pKa = 9.58NICFQCKK108 pKa = 6.91QCYY111 pKa = 8.9KK112 pKa = 10.83VIIEE116 pKa = 4.45SKK118 pKa = 8.72PVKK121 pKa = 10.15YY122 pKa = 10.52DD123 pKa = 3.4PEE125 pKa = 4.51VKK127 pKa = 10.39FFTLLLSNFQKK138 pKa = 10.87SSWRR142 pKa = 11.84RR143 pKa = 11.84SRR145 pKa = 11.84MCRR148 pKa = 11.84HH149 pKa = 6.36LEE151 pKa = 3.6WHH153 pKa = 6.47EE154 pKa = 4.01NEE156 pKa = 3.99QFSYY160 pKa = 10.86RR161 pKa = 11.84STRR164 pKa = 11.84PRR166 pKa = 11.84EE167 pKa = 4.12QGSEE171 pKa = 4.06PSPAATSSCSRR182 pKa = 11.84MDD184 pKa = 3.71SVPDD188 pKa = 3.76PPRR191 pKa = 11.84GPRR194 pKa = 11.84EE195 pKa = 4.06CLQGPSMSDD204 pKa = 3.3CASPIPGPCTSQEE217 pKa = 3.84PYY219 pKa = 11.19ADD221 pKa = 2.66VDD223 pKa = 3.73AFLEE227 pKa = 4.46GLLQQPQKK235 pKa = 11.0GRR237 pKa = 11.84ADD239 pKa = 3.21SGMGLDD245 pKa = 5.1PGAGWTEE252 pKa = 3.74DD253 pKa = 3.49LEE255 pKa = 4.86RR256 pKa = 11.84FILGDD261 pKa = 3.96DD262 pKa = 3.81NSCARR267 pKa = 11.84DD268 pKa = 3.42WTDD271 pKa = 4.09SMHH274 pKa = 7.16SDD276 pKa = 3.18SS277 pKa = 5.1
MM1 pKa = 7.52ASWNPGQKK9 pKa = 10.27GINIPQNDD17 pKa = 4.32EE18 pKa = 3.87SDD20 pKa = 3.86LPQPRR25 pKa = 11.84PLPALPASEE34 pKa = 5.13FMDD37 pKa = 3.58HH38 pKa = 6.77NYY40 pKa = 10.52VGMRR44 pKa = 11.84QWYY47 pKa = 8.32NDD49 pKa = 3.56PSGGYY54 pKa = 8.49PDD56 pKa = 4.9LGNEE60 pKa = 4.12EE61 pKa = 4.16LHH63 pKa = 7.43DD64 pKa = 3.98VLFSLAYY71 pKa = 10.16NCLHH75 pKa = 7.16FYY77 pKa = 10.7KK78 pKa = 10.26SLPIVRR84 pKa = 11.84QRR86 pKa = 11.84FKK88 pKa = 11.1CYY90 pKa = 10.32LEE92 pKa = 4.56PSFQRR97 pKa = 11.84VGIYY101 pKa = 9.58NICFQCKK108 pKa = 6.91QCYY111 pKa = 8.9KK112 pKa = 10.83VIIEE116 pKa = 4.45SKK118 pKa = 8.72PVKK121 pKa = 10.15YY122 pKa = 10.52DD123 pKa = 3.4PEE125 pKa = 4.51VKK127 pKa = 10.39FFTLLLSNFQKK138 pKa = 10.87SSWRR142 pKa = 11.84RR143 pKa = 11.84SRR145 pKa = 11.84MCRR148 pKa = 11.84HH149 pKa = 6.36LEE151 pKa = 3.6WHH153 pKa = 6.47EE154 pKa = 4.01NEE156 pKa = 3.99QFSYY160 pKa = 10.86RR161 pKa = 11.84STRR164 pKa = 11.84PRR166 pKa = 11.84EE167 pKa = 4.12QGSEE171 pKa = 4.06PSPAATSSCSRR182 pKa = 11.84MDD184 pKa = 3.71SVPDD188 pKa = 3.76PPRR191 pKa = 11.84GPRR194 pKa = 11.84EE195 pKa = 4.06CLQGPSMSDD204 pKa = 3.3CASPIPGPCTSQEE217 pKa = 3.84PYY219 pKa = 11.19ADD221 pKa = 2.66VDD223 pKa = 3.73AFLEE227 pKa = 4.46GLLQQPQKK235 pKa = 11.0GRR237 pKa = 11.84ADD239 pKa = 3.21SGMGLDD245 pKa = 5.1PGAGWTEE252 pKa = 3.74DD253 pKa = 3.49LEE255 pKa = 4.86RR256 pKa = 11.84FILGDD261 pKa = 3.96DD262 pKa = 3.81NSCARR267 pKa = 11.84DD268 pKa = 3.42WTDD271 pKa = 4.09SMHH274 pKa = 7.16SDD276 pKa = 3.18SS277 pKa = 5.1
Molecular weight: 31.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A088FA16|A0A088FA16_9RETR Bel1 protein OS=Brown greater galago prosimian foamy virus OX=2170139 GN=bel1 PE=4 SV=1
MM1 pKa = 7.12SQPSASGSAGAGGAPQQPPPPPPQPGPAAPVPRR34 pKa = 11.84AQIGYY39 pKa = 10.23GDD41 pKa = 5.11LDD43 pKa = 3.66VLLLQQEE50 pKa = 4.39YY51 pKa = 11.06HH52 pKa = 7.03LIDD55 pKa = 4.68PNLQVQHH62 pKa = 7.28LDD64 pKa = 3.29TLLVRR69 pKa = 11.84ITGGNWGPGDD79 pKa = 3.39RR80 pKa = 11.84FARR83 pKa = 11.84IEE85 pKa = 3.99VLLRR89 pKa = 11.84DD90 pKa = 3.85TLGPLQQPRR99 pKa = 11.84YY100 pKa = 9.27RR101 pKa = 11.84YY102 pKa = 10.11AAMQQADD109 pKa = 4.2LRR111 pKa = 11.84NDD113 pKa = 3.73IILHH117 pKa = 6.43LNYY120 pKa = 9.76QDD122 pKa = 4.81AIIIFDD128 pKa = 3.99MIIPSEE134 pKa = 4.19GVHH137 pKa = 5.14RR138 pKa = 11.84HH139 pKa = 4.91GPMFDD144 pKa = 3.68GLWIHH149 pKa = 7.07GDD151 pKa = 3.62DD152 pKa = 3.83YY153 pKa = 12.04SMNFQPITAHH163 pKa = 6.32EE164 pKa = 4.99LYY166 pKa = 10.82LLPQQVLTEE175 pKa = 4.08EE176 pKa = 4.54VEE178 pKa = 4.71LLTEE182 pKa = 4.1VCNRR186 pKa = 11.84MADD189 pKa = 3.75WIRR192 pKa = 11.84RR193 pKa = 11.84HH194 pKa = 5.65RR195 pKa = 11.84CGGGSGSSQPPPPPPPAVPVLPSAPPASSLPLPPQGWGISPPVATSTPGAAGHH248 pKa = 6.27SSSAGPNISLGGTYY262 pKa = 9.93VPPPVAPPAPVIGGPGGPGQLPAMVQVLPAQPVVIPINVIRR303 pKa = 11.84SVCGDD308 pKa = 3.68TPSNPQDD315 pKa = 3.03IPLWMGRR322 pKa = 11.84IIPAIEE328 pKa = 4.16GVFPIDD334 pKa = 3.81NPNLRR339 pKa = 11.84MRR341 pKa = 11.84VVNALLALHH350 pKa = 7.04PGLAITEE357 pKa = 4.57LNAQTWGQVLAVLHH371 pKa = 5.69MRR373 pKa = 11.84ALGHH377 pKa = 5.99TALHH381 pKa = 5.63QLPALLEE388 pKa = 4.56TIVKK392 pKa = 9.24TDD394 pKa = 4.56GILPAYY400 pKa = 9.49NMGMEE405 pKa = 4.49VTQQDD410 pKa = 3.94FSYY413 pKa = 10.79VWGILRR419 pKa = 11.84TLLPGQAFVLSMQNEE434 pKa = 4.76LDD436 pKa = 3.81RR437 pKa = 11.84LPAAQRR443 pKa = 11.84PGMFPGLLQRR453 pKa = 11.84TLDD456 pKa = 3.58ILGLNSRR463 pKa = 11.84GQNIQKK469 pKa = 9.26TNTQQQAPKK478 pKa = 9.87RR479 pKa = 11.84GQKK482 pKa = 9.44PKK484 pKa = 9.98PRR486 pKa = 11.84LPPVHH491 pKa = 6.87RR492 pKa = 11.84RR493 pKa = 11.84PAPFTPPATPSPRR506 pKa = 11.84QQASASPSSQGDD518 pKa = 3.28NRR520 pKa = 11.84SPQPQGRR527 pKa = 11.84GTYY530 pKa = 9.67GPSRR534 pKa = 11.84GGGSGPRR541 pKa = 11.84YY542 pKa = 9.0NFRR545 pKa = 11.84PRR547 pKa = 11.84VQPPDD552 pKa = 3.03RR553 pKa = 11.84YY554 pKa = 10.92GFGRR558 pKa = 11.84GQGGRR563 pKa = 11.84SSIGAQDD570 pKa = 3.58NQQPGQGGQRR580 pKa = 11.84TQQTNQNRR588 pKa = 11.84NQGNATGGRR597 pKa = 11.84TQPQNRR603 pKa = 11.84TVNTVRR609 pKa = 11.84VTQTNPQGGSSVSNPAVTTSQNTGTGSATQSSSSS643 pKa = 3.26
MM1 pKa = 7.12SQPSASGSAGAGGAPQQPPPPPPQPGPAAPVPRR34 pKa = 11.84AQIGYY39 pKa = 10.23GDD41 pKa = 5.11LDD43 pKa = 3.66VLLLQQEE50 pKa = 4.39YY51 pKa = 11.06HH52 pKa = 7.03LIDD55 pKa = 4.68PNLQVQHH62 pKa = 7.28LDD64 pKa = 3.29TLLVRR69 pKa = 11.84ITGGNWGPGDD79 pKa = 3.39RR80 pKa = 11.84FARR83 pKa = 11.84IEE85 pKa = 3.99VLLRR89 pKa = 11.84DD90 pKa = 3.85TLGPLQQPRR99 pKa = 11.84YY100 pKa = 9.27RR101 pKa = 11.84YY102 pKa = 10.11AAMQQADD109 pKa = 4.2LRR111 pKa = 11.84NDD113 pKa = 3.73IILHH117 pKa = 6.43LNYY120 pKa = 9.76QDD122 pKa = 4.81AIIIFDD128 pKa = 3.99MIIPSEE134 pKa = 4.19GVHH137 pKa = 5.14RR138 pKa = 11.84HH139 pKa = 4.91GPMFDD144 pKa = 3.68GLWIHH149 pKa = 7.07GDD151 pKa = 3.62DD152 pKa = 3.83YY153 pKa = 12.04SMNFQPITAHH163 pKa = 6.32EE164 pKa = 4.99LYY166 pKa = 10.82LLPQQVLTEE175 pKa = 4.08EE176 pKa = 4.54VEE178 pKa = 4.71LLTEE182 pKa = 4.1VCNRR186 pKa = 11.84MADD189 pKa = 3.75WIRR192 pKa = 11.84RR193 pKa = 11.84HH194 pKa = 5.65RR195 pKa = 11.84CGGGSGSSQPPPPPPPAVPVLPSAPPASSLPLPPQGWGISPPVATSTPGAAGHH248 pKa = 6.27SSSAGPNISLGGTYY262 pKa = 9.93VPPPVAPPAPVIGGPGGPGQLPAMVQVLPAQPVVIPINVIRR303 pKa = 11.84SVCGDD308 pKa = 3.68TPSNPQDD315 pKa = 3.03IPLWMGRR322 pKa = 11.84IIPAIEE328 pKa = 4.16GVFPIDD334 pKa = 3.81NPNLRR339 pKa = 11.84MRR341 pKa = 11.84VVNALLALHH350 pKa = 7.04PGLAITEE357 pKa = 4.57LNAQTWGQVLAVLHH371 pKa = 5.69MRR373 pKa = 11.84ALGHH377 pKa = 5.99TALHH381 pKa = 5.63QLPALLEE388 pKa = 4.56TIVKK392 pKa = 9.24TDD394 pKa = 4.56GILPAYY400 pKa = 9.49NMGMEE405 pKa = 4.49VTQQDD410 pKa = 3.94FSYY413 pKa = 10.79VWGILRR419 pKa = 11.84TLLPGQAFVLSMQNEE434 pKa = 4.76LDD436 pKa = 3.81RR437 pKa = 11.84LPAAQRR443 pKa = 11.84PGMFPGLLQRR453 pKa = 11.84TLDD456 pKa = 3.58ILGLNSRR463 pKa = 11.84GQNIQKK469 pKa = 9.26TNTQQQAPKK478 pKa = 9.87RR479 pKa = 11.84GQKK482 pKa = 9.44PKK484 pKa = 9.98PRR486 pKa = 11.84LPPVHH491 pKa = 6.87RR492 pKa = 11.84RR493 pKa = 11.84PAPFTPPATPSPRR506 pKa = 11.84QQASASPSSQGDD518 pKa = 3.28NRR520 pKa = 11.84SPQPQGRR527 pKa = 11.84GTYY530 pKa = 9.67GPSRR534 pKa = 11.84GGGSGPRR541 pKa = 11.84YY542 pKa = 9.0NFRR545 pKa = 11.84PRR547 pKa = 11.84VQPPDD552 pKa = 3.03RR553 pKa = 11.84YY554 pKa = 10.92GFGRR558 pKa = 11.84GQGGRR563 pKa = 11.84SSIGAQDD570 pKa = 3.58NQQPGQGGQRR580 pKa = 11.84TQQTNQNRR588 pKa = 11.84NQGNATGGRR597 pKa = 11.84TQPQNRR603 pKa = 11.84TVNTVRR609 pKa = 11.84VTQTNPQGGSSVSNPAVTTSQNTGTGSATQSSSSS643 pKa = 3.26
Molecular weight: 68.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2381 |
277 |
999 |
595.3 |
66.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.628 ± 0.564 | 2.1 ± 0.558 |
4.872 ± 0.507 | 4.914 ± 0.996 |
2.898 ± 0.426 | 6.846 ± 1.475 |
2.604 ± 0.231 | 5.25 ± 0.857 |
3.276 ± 1.006 | 9.534 ± 0.977 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.52 ± 0.127 | 4.326 ± 0.308 |
9.408 ± 2.067 | 6.216 ± 1.071 |
5.712 ± 0.465 | 7.224 ± 0.758 |
5.922 ± 0.936 | 5.502 ± 0.44 |
2.058 ± 0.397 | 3.192 ± 0.488 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
