
Capybara microvirus Cap3_SP_367
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.44
Get precalculated fractions of proteins
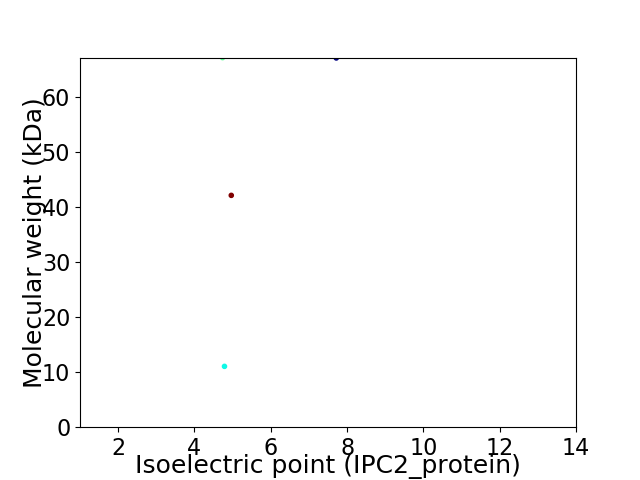
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A4P8W4U3|A0A4P8W4U3_9VIRU Replication initiator protein OS=Capybara microvirus Cap3_SP_367 OX=2585436 PE=4 SV=1
MM1 pKa = 7.69ANGFDD6 pKa = 4.04SLHH9 pKa = 6.08ITDD12 pKa = 4.88AVPNKK17 pKa = 10.66SKK19 pKa = 10.91FDD21 pKa = 3.73LSRR24 pKa = 11.84PHH26 pKa = 6.43LTTMDD31 pKa = 3.67FGEE34 pKa = 4.25ITPVFCEE41 pKa = 4.01EE42 pKa = 4.56GLPGDD47 pKa = 4.21KK48 pKa = 9.92FTVDD52 pKa = 2.99MTAFARR58 pKa = 11.84LAPMVFPTYY67 pKa = 10.61GKK69 pKa = 10.45CCLKK73 pKa = 10.66SAAYY77 pKa = 9.91YY78 pKa = 9.58VDD80 pKa = 4.25YY81 pKa = 11.0YY82 pKa = 11.53QLADD86 pKa = 3.98DD87 pKa = 5.29FEE89 pKa = 6.05AFISGQTTYY98 pKa = 11.04RR99 pKa = 11.84GTTPQLRR106 pKa = 11.84VISADD111 pKa = 3.92GILDD115 pKa = 4.49LLTSNPSSEE124 pKa = 4.11FFNDD128 pKa = 3.4LSTEE132 pKa = 3.76IANPSSEE139 pKa = 4.18SEE141 pKa = 3.92YY142 pKa = 11.23DD143 pKa = 3.26FTFDD147 pKa = 3.15EE148 pKa = 5.25DD149 pKa = 5.55GIILYY154 pKa = 10.41YY155 pKa = 10.66KK156 pKa = 8.59FTRR159 pKa = 11.84KK160 pKa = 9.6GRR162 pKa = 11.84WVFKK166 pKa = 9.94CLRR169 pKa = 11.84SLGYY173 pKa = 10.15DD174 pKa = 2.83IPTQRR179 pKa = 11.84GDD181 pKa = 3.35SSVFSAMPLLAFFKK195 pKa = 10.61AYY197 pKa = 10.31NDD199 pKa = 3.55YY200 pKa = 10.0MALNSIQNMNITSQMLQAIKK220 pKa = 10.67NKK222 pKa = 9.24VSASVTVYY230 pKa = 9.99PYY232 pKa = 10.71EE233 pKa = 5.23GGAGSEE239 pKa = 5.01GYY241 pKa = 7.87TVSYY245 pKa = 10.41NGSNVISGDD254 pKa = 3.52VIRR257 pKa = 11.84AMFDD261 pKa = 3.76DD262 pKa = 4.75IRR264 pKa = 11.84LCFDD268 pKa = 3.23NDD270 pKa = 3.72YY271 pKa = 9.93VTSLWSTANQAYY283 pKa = 9.2PGLSQNITTPTLNGVQMNQSVVKK306 pKa = 10.03DD307 pKa = 3.55WNNTSVSGISGSISQRR323 pKa = 11.84SLDD326 pKa = 3.81VLRR329 pKa = 11.84MFDD332 pKa = 2.72NWVRR336 pKa = 11.84RR337 pKa = 11.84NNYY340 pKa = 9.41VGSKK344 pKa = 10.15DD345 pKa = 3.46VNQIYY350 pKa = 10.68ARR352 pKa = 11.84FGIHH356 pKa = 6.95PEE358 pKa = 3.81NFKK361 pKa = 11.09AKK363 pKa = 8.03YY364 pKa = 9.34SHH366 pKa = 6.86MIGKK370 pKa = 9.16SEE372 pKa = 4.48DD373 pKa = 3.86CIQVGDD379 pKa = 3.87VTSTSDD385 pKa = 3.26TYY387 pKa = 11.59QDD389 pKa = 3.35ASNGAVIGSYY399 pKa = 9.82AGKK402 pKa = 10.45GIASGDD408 pKa = 3.77SKK410 pKa = 10.89FVYY413 pKa = 10.58SPDD416 pKa = 3.31EE417 pKa = 4.29FGCLIILSWISVRR430 pKa = 11.84PMYY433 pKa = 10.28TRR435 pKa = 11.84GFQRR439 pKa = 11.84HH440 pKa = 5.61TLRR443 pKa = 11.84QSPTDD448 pKa = 4.11FYY450 pKa = 11.03TPEE453 pKa = 3.92FDD455 pKa = 4.09GVGGNAVSYY464 pKa = 11.15RR465 pKa = 11.84EE466 pKa = 3.74ITVPKK471 pKa = 9.7TVDD474 pKa = 3.67DD475 pKa = 4.31EE476 pKa = 5.36SMGYY480 pKa = 10.51YY481 pKa = 10.13NDD483 pKa = 3.49GVGFNEE489 pKa = 4.97RR490 pKa = 11.84YY491 pKa = 9.67CEE493 pKa = 3.92YY494 pKa = 10.43RR495 pKa = 11.84DD496 pKa = 3.61SSKK499 pKa = 11.07KK500 pKa = 10.83GIVSGDD506 pKa = 3.5FLLDD510 pKa = 3.13TGMDD514 pKa = 2.96AWHH517 pKa = 7.23FGRR520 pKa = 11.84DD521 pKa = 3.48LDD523 pKa = 5.11SLYY526 pKa = 10.65TSHH529 pKa = 7.19NLGAQTYY536 pKa = 9.4ALNTYY541 pKa = 7.92GVPGSVSEE549 pKa = 4.13YY550 pKa = 11.22DD551 pKa = 4.08RR552 pKa = 11.84IFNNAGSVAGRR563 pKa = 11.84DD564 pKa = 3.22HH565 pKa = 6.91FFISFYY571 pKa = 11.05FNVQAIRR578 pKa = 11.84PIKK581 pKa = 10.4NMNAVADD588 pKa = 4.59LGSGDD593 pKa = 3.64INISKK598 pKa = 10.36VGNEE602 pKa = 4.03VNN604 pKa = 3.5
MM1 pKa = 7.69ANGFDD6 pKa = 4.04SLHH9 pKa = 6.08ITDD12 pKa = 4.88AVPNKK17 pKa = 10.66SKK19 pKa = 10.91FDD21 pKa = 3.73LSRR24 pKa = 11.84PHH26 pKa = 6.43LTTMDD31 pKa = 3.67FGEE34 pKa = 4.25ITPVFCEE41 pKa = 4.01EE42 pKa = 4.56GLPGDD47 pKa = 4.21KK48 pKa = 9.92FTVDD52 pKa = 2.99MTAFARR58 pKa = 11.84LAPMVFPTYY67 pKa = 10.61GKK69 pKa = 10.45CCLKK73 pKa = 10.66SAAYY77 pKa = 9.91YY78 pKa = 9.58VDD80 pKa = 4.25YY81 pKa = 11.0YY82 pKa = 11.53QLADD86 pKa = 3.98DD87 pKa = 5.29FEE89 pKa = 6.05AFISGQTTYY98 pKa = 11.04RR99 pKa = 11.84GTTPQLRR106 pKa = 11.84VISADD111 pKa = 3.92GILDD115 pKa = 4.49LLTSNPSSEE124 pKa = 4.11FFNDD128 pKa = 3.4LSTEE132 pKa = 3.76IANPSSEE139 pKa = 4.18SEE141 pKa = 3.92YY142 pKa = 11.23DD143 pKa = 3.26FTFDD147 pKa = 3.15EE148 pKa = 5.25DD149 pKa = 5.55GIILYY154 pKa = 10.41YY155 pKa = 10.66KK156 pKa = 8.59FTRR159 pKa = 11.84KK160 pKa = 9.6GRR162 pKa = 11.84WVFKK166 pKa = 9.94CLRR169 pKa = 11.84SLGYY173 pKa = 10.15DD174 pKa = 2.83IPTQRR179 pKa = 11.84GDD181 pKa = 3.35SSVFSAMPLLAFFKK195 pKa = 10.61AYY197 pKa = 10.31NDD199 pKa = 3.55YY200 pKa = 10.0MALNSIQNMNITSQMLQAIKK220 pKa = 10.67NKK222 pKa = 9.24VSASVTVYY230 pKa = 9.99PYY232 pKa = 10.71EE233 pKa = 5.23GGAGSEE239 pKa = 5.01GYY241 pKa = 7.87TVSYY245 pKa = 10.41NGSNVISGDD254 pKa = 3.52VIRR257 pKa = 11.84AMFDD261 pKa = 3.76DD262 pKa = 4.75IRR264 pKa = 11.84LCFDD268 pKa = 3.23NDD270 pKa = 3.72YY271 pKa = 9.93VTSLWSTANQAYY283 pKa = 9.2PGLSQNITTPTLNGVQMNQSVVKK306 pKa = 10.03DD307 pKa = 3.55WNNTSVSGISGSISQRR323 pKa = 11.84SLDD326 pKa = 3.81VLRR329 pKa = 11.84MFDD332 pKa = 2.72NWVRR336 pKa = 11.84RR337 pKa = 11.84NNYY340 pKa = 9.41VGSKK344 pKa = 10.15DD345 pKa = 3.46VNQIYY350 pKa = 10.68ARR352 pKa = 11.84FGIHH356 pKa = 6.95PEE358 pKa = 3.81NFKK361 pKa = 11.09AKK363 pKa = 8.03YY364 pKa = 9.34SHH366 pKa = 6.86MIGKK370 pKa = 9.16SEE372 pKa = 4.48DD373 pKa = 3.86CIQVGDD379 pKa = 3.87VTSTSDD385 pKa = 3.26TYY387 pKa = 11.59QDD389 pKa = 3.35ASNGAVIGSYY399 pKa = 9.82AGKK402 pKa = 10.45GIASGDD408 pKa = 3.77SKK410 pKa = 10.89FVYY413 pKa = 10.58SPDD416 pKa = 3.31EE417 pKa = 4.29FGCLIILSWISVRR430 pKa = 11.84PMYY433 pKa = 10.28TRR435 pKa = 11.84GFQRR439 pKa = 11.84HH440 pKa = 5.61TLRR443 pKa = 11.84QSPTDD448 pKa = 4.11FYY450 pKa = 11.03TPEE453 pKa = 3.92FDD455 pKa = 4.09GVGGNAVSYY464 pKa = 11.15RR465 pKa = 11.84EE466 pKa = 3.74ITVPKK471 pKa = 9.7TVDD474 pKa = 3.67DD475 pKa = 4.31EE476 pKa = 5.36SMGYY480 pKa = 10.51YY481 pKa = 10.13NDD483 pKa = 3.49GVGFNEE489 pKa = 4.97RR490 pKa = 11.84YY491 pKa = 9.67CEE493 pKa = 3.92YY494 pKa = 10.43RR495 pKa = 11.84DD496 pKa = 3.61SSKK499 pKa = 11.07KK500 pKa = 10.83GIVSGDD506 pKa = 3.5FLLDD510 pKa = 3.13TGMDD514 pKa = 2.96AWHH517 pKa = 7.23FGRR520 pKa = 11.84DD521 pKa = 3.48LDD523 pKa = 5.11SLYY526 pKa = 10.65TSHH529 pKa = 7.19NLGAQTYY536 pKa = 9.4ALNTYY541 pKa = 7.92GVPGSVSEE549 pKa = 4.13YY550 pKa = 11.22DD551 pKa = 4.08RR552 pKa = 11.84IFNNAGSVAGRR563 pKa = 11.84DD564 pKa = 3.22HH565 pKa = 6.91FFISFYY571 pKa = 11.05FNVQAIRR578 pKa = 11.84PIKK581 pKa = 10.4NMNAVADD588 pKa = 4.59LGSGDD593 pKa = 3.64INISKK598 pKa = 10.36VGNEE602 pKa = 4.03VNN604 pKa = 3.5
Molecular weight: 67.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W584|A0A4P8W584_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_367 OX=2585436 PE=4 SV=1
MM1 pKa = 8.47DD2 pKa = 4.71ILNRR6 pKa = 11.84NKK8 pKa = 10.48NFIPGKK14 pKa = 10.21DD15 pKa = 2.96KK16 pKa = 10.93VTFSLPDD23 pKa = 3.4MKK25 pKa = 10.89SYY27 pKa = 10.32IYY29 pKa = 9.6SQRR32 pKa = 11.84KK33 pKa = 7.08IEE35 pKa = 4.13NYY37 pKa = 7.38EE38 pKa = 3.81TLAYY42 pKa = 8.62YY43 pKa = 8.79QWKK46 pKa = 8.1EE47 pKa = 3.92TVDD50 pKa = 4.06NDD52 pKa = 4.1GQCYY56 pKa = 9.4FYY58 pKa = 10.91TLTYY62 pKa = 10.49NDD64 pKa = 3.18NALPKK69 pKa = 10.29FNGVPCFDD77 pKa = 4.75YY78 pKa = 11.04EE79 pKa = 4.47DD80 pKa = 3.81HH81 pKa = 8.1DD82 pKa = 4.32KK83 pKa = 11.14FLDD86 pKa = 3.83VLSHH90 pKa = 6.87KK91 pKa = 10.38LLRR94 pKa = 11.84SYY96 pKa = 8.75GTKK99 pKa = 10.32LKK101 pKa = 10.8YY102 pKa = 10.52FSTTEE107 pKa = 3.71RR108 pKa = 11.84GEE110 pKa = 4.28GKK112 pKa = 10.49GFRR115 pKa = 11.84GKK117 pKa = 10.79GNNPHH122 pKa = 5.14YY123 pKa = 11.09HH124 pKa = 5.78MLFFLTNANNPNYY137 pKa = 9.46SYY139 pKa = 11.53KK140 pKa = 10.64KK141 pKa = 9.81IRR143 pKa = 11.84PTDD146 pKa = 3.7FCAIVKK152 pKa = 9.61NYY154 pKa = 8.0WQGTTKK160 pKa = 10.38EE161 pKa = 4.31GKK163 pKa = 9.55NVAQHH168 pKa = 5.31YY169 pKa = 9.35KK170 pKa = 10.6YY171 pKa = 10.79GIAAPGDD178 pKa = 3.38NFGLVKK184 pKa = 10.54SPQAVTYY191 pKa = 9.27CCKK194 pKa = 10.43YY195 pKa = 8.1VCKK198 pKa = 10.5DD199 pKa = 2.79AAEE202 pKa = 4.13RR203 pKa = 11.84SNYY206 pKa = 9.79DD207 pKa = 3.25SQVHH211 pKa = 5.65YY212 pKa = 10.5IVSKK216 pKa = 9.58TINEE220 pKa = 4.41TLSSVHH226 pKa = 5.66IQEE229 pKa = 4.64EE230 pKa = 4.47FLLSQVFSRR239 pKa = 11.84DD240 pKa = 3.26GLSMKK245 pKa = 10.3EE246 pKa = 4.03GLSMIKK252 pKa = 10.1DD253 pKa = 3.61CYY255 pKa = 10.94LRR257 pKa = 11.84TEE259 pKa = 4.45INSKK263 pKa = 8.88DD264 pKa = 3.63TPSTISYY271 pKa = 8.89WFIIHH276 pKa = 6.62SGLYY280 pKa = 10.32DD281 pKa = 3.55EE282 pKa = 5.19FRR284 pKa = 11.84SYY286 pKa = 11.13CILEE290 pKa = 4.4CQDD293 pKa = 4.0KK294 pKa = 10.58IRR296 pKa = 11.84EE297 pKa = 4.15RR298 pKa = 11.84VNEE301 pKa = 3.61FRR303 pKa = 11.84RR304 pKa = 11.84RR305 pKa = 11.84YY306 pKa = 8.23CGKK309 pKa = 9.57VRR311 pKa = 11.84HH312 pKa = 6.45SNDD315 pKa = 2.9FGLSGLSTIKK325 pKa = 10.83DD326 pKa = 3.56DD327 pKa = 3.58MNPRR331 pKa = 11.84VLFPSKK337 pKa = 10.18KK338 pKa = 8.35GYY340 pKa = 10.15KK341 pKa = 9.42YY342 pKa = 10.51RR343 pKa = 11.84PLPLYY348 pKa = 10.03YY349 pKa = 9.94YY350 pKa = 9.55RR351 pKa = 11.84KK352 pKa = 9.05KK353 pKa = 10.57YY354 pKa = 10.48CVVDD358 pKa = 3.37YY359 pKa = 11.34YY360 pKa = 10.95MQGDD364 pKa = 3.83KK365 pKa = 10.7KK366 pKa = 10.61IPYY369 pKa = 9.22YY370 pKa = 10.45RR371 pKa = 11.84LNDD374 pKa = 3.83LGVKK378 pKa = 8.03YY379 pKa = 9.37TNFKK383 pKa = 10.5LDD385 pKa = 3.45SLIDD389 pKa = 3.72SKK391 pKa = 11.48KK392 pKa = 8.81ITTEE396 pKa = 3.66KK397 pKa = 10.87ALSFVDD403 pKa = 4.01KK404 pKa = 10.93LGSGYY409 pKa = 10.27FYY411 pKa = 11.17SEE413 pKa = 4.02QFKK416 pKa = 11.11SLGMPEE422 pKa = 4.66NKK424 pKa = 9.34VKK426 pKa = 10.96YY427 pKa = 10.56LIGDD431 pKa = 4.36LNSSIKK437 pKa = 10.56DD438 pKa = 3.43NYY440 pKa = 9.77SVYY443 pKa = 10.88SVVYY447 pKa = 10.14KK448 pKa = 10.76NRR450 pKa = 11.84SLSGLEE456 pKa = 4.97DD457 pKa = 3.28IDD459 pKa = 5.73RR460 pKa = 11.84PLDD463 pKa = 3.68MYY465 pKa = 11.69NDD467 pKa = 3.01WCYY470 pKa = 11.3FNTPVIKK477 pKa = 10.2EE478 pKa = 4.37DD479 pKa = 3.36NSNTYY484 pKa = 8.04TAYY487 pKa = 10.7AFLKK491 pKa = 10.73SFTRR495 pKa = 11.84YY496 pKa = 10.74DD497 pKa = 3.44FTDD500 pKa = 4.19DD501 pKa = 4.29GLVHH505 pKa = 6.44TNSLTYY511 pKa = 9.93RR512 pKa = 11.84YY513 pKa = 9.54HH514 pKa = 7.78KK515 pKa = 10.76DD516 pKa = 3.3FEE518 pKa = 4.66CFLEE522 pKa = 4.53YY523 pKa = 10.42FDD525 pKa = 5.93LLDD528 pKa = 4.97SIIDD532 pKa = 3.83YY533 pKa = 11.39YY534 pKa = 11.0VMCDD538 pKa = 3.24SLEE541 pKa = 4.16KK542 pKa = 10.93EE543 pKa = 4.11NIARR547 pKa = 11.84QWRR550 pKa = 11.84DD551 pKa = 2.71TRR553 pKa = 11.84NVINSIKK560 pKa = 10.17HH561 pKa = 4.89PEE563 pKa = 4.1YY564 pKa = 10.85YY565 pKa = 10.79GLAVV569 pKa = 3.44
MM1 pKa = 8.47DD2 pKa = 4.71ILNRR6 pKa = 11.84NKK8 pKa = 10.48NFIPGKK14 pKa = 10.21DD15 pKa = 2.96KK16 pKa = 10.93VTFSLPDD23 pKa = 3.4MKK25 pKa = 10.89SYY27 pKa = 10.32IYY29 pKa = 9.6SQRR32 pKa = 11.84KK33 pKa = 7.08IEE35 pKa = 4.13NYY37 pKa = 7.38EE38 pKa = 3.81TLAYY42 pKa = 8.62YY43 pKa = 8.79QWKK46 pKa = 8.1EE47 pKa = 3.92TVDD50 pKa = 4.06NDD52 pKa = 4.1GQCYY56 pKa = 9.4FYY58 pKa = 10.91TLTYY62 pKa = 10.49NDD64 pKa = 3.18NALPKK69 pKa = 10.29FNGVPCFDD77 pKa = 4.75YY78 pKa = 11.04EE79 pKa = 4.47DD80 pKa = 3.81HH81 pKa = 8.1DD82 pKa = 4.32KK83 pKa = 11.14FLDD86 pKa = 3.83VLSHH90 pKa = 6.87KK91 pKa = 10.38LLRR94 pKa = 11.84SYY96 pKa = 8.75GTKK99 pKa = 10.32LKK101 pKa = 10.8YY102 pKa = 10.52FSTTEE107 pKa = 3.71RR108 pKa = 11.84GEE110 pKa = 4.28GKK112 pKa = 10.49GFRR115 pKa = 11.84GKK117 pKa = 10.79GNNPHH122 pKa = 5.14YY123 pKa = 11.09HH124 pKa = 5.78MLFFLTNANNPNYY137 pKa = 9.46SYY139 pKa = 11.53KK140 pKa = 10.64KK141 pKa = 9.81IRR143 pKa = 11.84PTDD146 pKa = 3.7FCAIVKK152 pKa = 9.61NYY154 pKa = 8.0WQGTTKK160 pKa = 10.38EE161 pKa = 4.31GKK163 pKa = 9.55NVAQHH168 pKa = 5.31YY169 pKa = 9.35KK170 pKa = 10.6YY171 pKa = 10.79GIAAPGDD178 pKa = 3.38NFGLVKK184 pKa = 10.54SPQAVTYY191 pKa = 9.27CCKK194 pKa = 10.43YY195 pKa = 8.1VCKK198 pKa = 10.5DD199 pKa = 2.79AAEE202 pKa = 4.13RR203 pKa = 11.84SNYY206 pKa = 9.79DD207 pKa = 3.25SQVHH211 pKa = 5.65YY212 pKa = 10.5IVSKK216 pKa = 9.58TINEE220 pKa = 4.41TLSSVHH226 pKa = 5.66IQEE229 pKa = 4.64EE230 pKa = 4.47FLLSQVFSRR239 pKa = 11.84DD240 pKa = 3.26GLSMKK245 pKa = 10.3EE246 pKa = 4.03GLSMIKK252 pKa = 10.1DD253 pKa = 3.61CYY255 pKa = 10.94LRR257 pKa = 11.84TEE259 pKa = 4.45INSKK263 pKa = 8.88DD264 pKa = 3.63TPSTISYY271 pKa = 8.89WFIIHH276 pKa = 6.62SGLYY280 pKa = 10.32DD281 pKa = 3.55EE282 pKa = 5.19FRR284 pKa = 11.84SYY286 pKa = 11.13CILEE290 pKa = 4.4CQDD293 pKa = 4.0KK294 pKa = 10.58IRR296 pKa = 11.84EE297 pKa = 4.15RR298 pKa = 11.84VNEE301 pKa = 3.61FRR303 pKa = 11.84RR304 pKa = 11.84RR305 pKa = 11.84YY306 pKa = 8.23CGKK309 pKa = 9.57VRR311 pKa = 11.84HH312 pKa = 6.45SNDD315 pKa = 2.9FGLSGLSTIKK325 pKa = 10.83DD326 pKa = 3.56DD327 pKa = 3.58MNPRR331 pKa = 11.84VLFPSKK337 pKa = 10.18KK338 pKa = 8.35GYY340 pKa = 10.15KK341 pKa = 9.42YY342 pKa = 10.51RR343 pKa = 11.84PLPLYY348 pKa = 10.03YY349 pKa = 9.94YY350 pKa = 9.55RR351 pKa = 11.84KK352 pKa = 9.05KK353 pKa = 10.57YY354 pKa = 10.48CVVDD358 pKa = 3.37YY359 pKa = 11.34YY360 pKa = 10.95MQGDD364 pKa = 3.83KK365 pKa = 10.7KK366 pKa = 10.61IPYY369 pKa = 9.22YY370 pKa = 10.45RR371 pKa = 11.84LNDD374 pKa = 3.83LGVKK378 pKa = 8.03YY379 pKa = 9.37TNFKK383 pKa = 10.5LDD385 pKa = 3.45SLIDD389 pKa = 3.72SKK391 pKa = 11.48KK392 pKa = 8.81ITTEE396 pKa = 3.66KK397 pKa = 10.87ALSFVDD403 pKa = 4.01KK404 pKa = 10.93LGSGYY409 pKa = 10.27FYY411 pKa = 11.17SEE413 pKa = 4.02QFKK416 pKa = 11.11SLGMPEE422 pKa = 4.66NKK424 pKa = 9.34VKK426 pKa = 10.96YY427 pKa = 10.56LIGDD431 pKa = 4.36LNSSIKK437 pKa = 10.56DD438 pKa = 3.43NYY440 pKa = 9.77SVYY443 pKa = 10.88SVVYY447 pKa = 10.14KK448 pKa = 10.76NRR450 pKa = 11.84SLSGLEE456 pKa = 4.97DD457 pKa = 3.28IDD459 pKa = 5.73RR460 pKa = 11.84PLDD463 pKa = 3.68MYY465 pKa = 11.69NDD467 pKa = 3.01WCYY470 pKa = 11.3FNTPVIKK477 pKa = 10.2EE478 pKa = 4.37DD479 pKa = 3.36NSNTYY484 pKa = 8.04TAYY487 pKa = 10.7AFLKK491 pKa = 10.73SFTRR495 pKa = 11.84YY496 pKa = 10.74DD497 pKa = 3.44FTDD500 pKa = 4.19DD501 pKa = 4.29GLVHH505 pKa = 6.44TNSLTYY511 pKa = 9.93RR512 pKa = 11.84YY513 pKa = 9.54HH514 pKa = 7.78KK515 pKa = 10.76DD516 pKa = 3.3FEE518 pKa = 4.66CFLEE522 pKa = 4.53YY523 pKa = 10.42FDD525 pKa = 5.93LLDD528 pKa = 4.97SIIDD532 pKa = 3.83YY533 pKa = 11.39YY534 pKa = 11.0VMCDD538 pKa = 3.24SLEE541 pKa = 4.16KK542 pKa = 10.93EE543 pKa = 4.11NIARR547 pKa = 11.84QWRR550 pKa = 11.84DD551 pKa = 2.71TRR553 pKa = 11.84NVINSIKK560 pKa = 10.17HH561 pKa = 4.89PEE563 pKa = 4.1YY564 pKa = 10.85YY565 pKa = 10.79GLAVV569 pKa = 3.44
Molecular weight: 67.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1648 |
98 |
604 |
412.0 |
46.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.886 ± 1.797 | 1.578 ± 0.381 |
7.464 ± 0.6 | 5.34 ± 1.151 |
4.672 ± 0.804 | 6.25 ± 0.918 |
1.335 ± 0.38 | 5.825 ± 0.323 |
6.978 ± 1.359 | 6.675 ± 0.601 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.367 ± 0.249 | 6.553 ± 0.33 |
3.459 ± 0.183 | 3.762 ± 0.984 |
4.126 ± 0.324 | 8.495 ± 0.862 |
5.4 ± 0.384 | 6.189 ± 0.447 |
1.032 ± 0.166 | 6.614 ± 1.362 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
