
Human papillomavirus 116
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 9
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
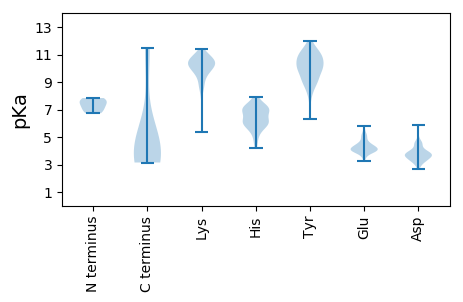
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|C7B7D8|C7B7D8_9PAPI Major capsid protein L1 OS=Human papillomavirus 116 OX=915428 GN=L1 PE=3 SV=1
MM1 pKa = 6.73MRR3 pKa = 11.84IRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.63RR8 pKa = 11.84ASPTDD13 pKa = 3.26LHH15 pKa = 6.82RR16 pKa = 11.84SCALGGDD23 pKa = 4.38CFPDD27 pKa = 3.63VEE29 pKa = 4.79NKK31 pKa = 10.71LSGNTLADD39 pKa = 3.31ILLKK43 pKa = 10.95AFGSILYY50 pKa = 9.92LGNLGIGTGKK60 pKa = 10.75GGGGQYY66 pKa = 10.87GYY68 pKa = 9.03TPFGGTRR75 pKa = 11.84PSISRR80 pKa = 11.84APVRR84 pKa = 11.84PAIPVDD90 pKa = 3.62TVVPGEE96 pKa = 4.18VLPVTPLDD104 pKa = 3.69PAIVPLTDD112 pKa = 4.11GLPEE116 pKa = 3.95PAVIDD121 pKa = 3.77VPGAGPGLPTEE132 pKa = 4.75TIDD135 pKa = 3.72VTTEE139 pKa = 3.56LDD141 pKa = 3.63LVSEE145 pKa = 4.36VTGVGEE151 pKa = 4.54HH152 pKa = 7.02PSVTYY157 pKa = 8.35DD158 pKa = 3.47TNNVAQIDD166 pKa = 4.35VQVQPPPPKK175 pKa = 10.18RR176 pKa = 11.84ILLDD180 pKa = 3.32TSITDD185 pKa = 3.27TEE187 pKa = 4.61LAVQTHH193 pKa = 6.81ASHH196 pKa = 7.32VDD198 pKa = 2.78EE199 pKa = 5.45HH200 pKa = 7.92YY201 pKa = 11.37NVFVDD206 pKa = 3.93AQFHH210 pKa = 6.37GEE212 pKa = 4.12HH213 pKa = 6.47IGAFEE218 pKa = 4.27PEE220 pKa = 4.47SIEE223 pKa = 3.85LQEE226 pKa = 3.94INLRR230 pKa = 11.84QEE232 pKa = 3.99FEE234 pKa = 3.89IDD236 pKa = 3.25EE237 pKa = 4.84GPLRR241 pKa = 11.84STPLSSRR248 pKa = 11.84AISRR252 pKa = 11.84ARR254 pKa = 11.84DD255 pKa = 3.24LYY257 pKa = 10.83NRR259 pKa = 11.84YY260 pKa = 8.37VQQVPTTQLASVSTPRR276 pKa = 11.84VTFEE280 pKa = 3.97FEE282 pKa = 3.88NPAFEE287 pKa = 5.61AEE289 pKa = 4.1IADD292 pKa = 3.68VFNRR296 pKa = 11.84EE297 pKa = 4.04VQEE300 pKa = 4.95LATQGQDD307 pKa = 2.95EE308 pKa = 5.19AGTDD312 pKa = 4.63LIRR315 pKa = 11.84LSDD318 pKa = 3.31IRR320 pKa = 11.84YY321 pKa = 8.73GEE323 pKa = 4.4SPAGTVRR330 pKa = 11.84VSRR333 pKa = 11.84LGQRR337 pKa = 11.84EE338 pKa = 4.1GMIMRR343 pKa = 11.84SGLQVGQRR351 pKa = 11.84VHH353 pKa = 7.1FYY355 pKa = 11.11YY356 pKa = 10.66DD357 pKa = 3.39ISPIPKK363 pKa = 9.48EE364 pKa = 4.18AIEE367 pKa = 4.04LRR369 pKa = 11.84TFGEE373 pKa = 4.34YY374 pKa = 8.98SHH376 pKa = 7.3EE377 pKa = 4.13YY378 pKa = 9.53TVVDD382 pKa = 4.66DD383 pKa = 4.6LASSSFINPFEE394 pKa = 4.11QPVDD398 pKa = 3.56GSLEE402 pKa = 3.94FSDD405 pKa = 4.65AALIDD410 pKa = 3.96SVEE413 pKa = 4.03EE414 pKa = 4.33DD415 pKa = 3.65FSGTHH420 pKa = 7.6LILTSANAADD430 pKa = 4.77SIDD433 pKa = 3.73IPVIPPGIGVRR444 pKa = 11.84VFVDD448 pKa = 4.99DD449 pKa = 3.83YY450 pKa = 12.01AKK452 pKa = 10.64GLSVLHH458 pKa = 5.91PTIIDD463 pKa = 3.3NGAIYY468 pKa = 8.15PTDD471 pKa = 3.22ISSNILPLTPSFSIDD486 pKa = 3.39VNYY489 pKa = 10.76SDD491 pKa = 5.76YY492 pKa = 11.17NIHH495 pKa = 6.93PANIRR500 pKa = 11.84RR501 pKa = 11.84KK502 pKa = 8.5RR503 pKa = 11.84KK504 pKa = 9.36HH505 pKa = 6.09SSSLYY510 pKa = 9.47FF511 pKa = 3.99
MM1 pKa = 6.73MRR3 pKa = 11.84IRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.63RR8 pKa = 11.84ASPTDD13 pKa = 3.26LHH15 pKa = 6.82RR16 pKa = 11.84SCALGGDD23 pKa = 4.38CFPDD27 pKa = 3.63VEE29 pKa = 4.79NKK31 pKa = 10.71LSGNTLADD39 pKa = 3.31ILLKK43 pKa = 10.95AFGSILYY50 pKa = 9.92LGNLGIGTGKK60 pKa = 10.75GGGGQYY66 pKa = 10.87GYY68 pKa = 9.03TPFGGTRR75 pKa = 11.84PSISRR80 pKa = 11.84APVRR84 pKa = 11.84PAIPVDD90 pKa = 3.62TVVPGEE96 pKa = 4.18VLPVTPLDD104 pKa = 3.69PAIVPLTDD112 pKa = 4.11GLPEE116 pKa = 3.95PAVIDD121 pKa = 3.77VPGAGPGLPTEE132 pKa = 4.75TIDD135 pKa = 3.72VTTEE139 pKa = 3.56LDD141 pKa = 3.63LVSEE145 pKa = 4.36VTGVGEE151 pKa = 4.54HH152 pKa = 7.02PSVTYY157 pKa = 8.35DD158 pKa = 3.47TNNVAQIDD166 pKa = 4.35VQVQPPPPKK175 pKa = 10.18RR176 pKa = 11.84ILLDD180 pKa = 3.32TSITDD185 pKa = 3.27TEE187 pKa = 4.61LAVQTHH193 pKa = 6.81ASHH196 pKa = 7.32VDD198 pKa = 2.78EE199 pKa = 5.45HH200 pKa = 7.92YY201 pKa = 11.37NVFVDD206 pKa = 3.93AQFHH210 pKa = 6.37GEE212 pKa = 4.12HH213 pKa = 6.47IGAFEE218 pKa = 4.27PEE220 pKa = 4.47SIEE223 pKa = 3.85LQEE226 pKa = 3.94INLRR230 pKa = 11.84QEE232 pKa = 3.99FEE234 pKa = 3.89IDD236 pKa = 3.25EE237 pKa = 4.84GPLRR241 pKa = 11.84STPLSSRR248 pKa = 11.84AISRR252 pKa = 11.84ARR254 pKa = 11.84DD255 pKa = 3.24LYY257 pKa = 10.83NRR259 pKa = 11.84YY260 pKa = 8.37VQQVPTTQLASVSTPRR276 pKa = 11.84VTFEE280 pKa = 3.97FEE282 pKa = 3.88NPAFEE287 pKa = 5.61AEE289 pKa = 4.1IADD292 pKa = 3.68VFNRR296 pKa = 11.84EE297 pKa = 4.04VQEE300 pKa = 4.95LATQGQDD307 pKa = 2.95EE308 pKa = 5.19AGTDD312 pKa = 4.63LIRR315 pKa = 11.84LSDD318 pKa = 3.31IRR320 pKa = 11.84YY321 pKa = 8.73GEE323 pKa = 4.4SPAGTVRR330 pKa = 11.84VSRR333 pKa = 11.84LGQRR337 pKa = 11.84EE338 pKa = 4.1GMIMRR343 pKa = 11.84SGLQVGQRR351 pKa = 11.84VHH353 pKa = 7.1FYY355 pKa = 11.11YY356 pKa = 10.66DD357 pKa = 3.39ISPIPKK363 pKa = 9.48EE364 pKa = 4.18AIEE367 pKa = 4.04LRR369 pKa = 11.84TFGEE373 pKa = 4.34YY374 pKa = 8.98SHH376 pKa = 7.3EE377 pKa = 4.13YY378 pKa = 9.53TVVDD382 pKa = 4.66DD383 pKa = 4.6LASSSFINPFEE394 pKa = 4.11QPVDD398 pKa = 3.56GSLEE402 pKa = 3.94FSDD405 pKa = 4.65AALIDD410 pKa = 3.96SVEE413 pKa = 4.03EE414 pKa = 4.33DD415 pKa = 3.65FSGTHH420 pKa = 7.6LILTSANAADD430 pKa = 4.77SIDD433 pKa = 3.73IPVIPPGIGVRR444 pKa = 11.84VFVDD448 pKa = 4.99DD449 pKa = 3.83YY450 pKa = 12.01AKK452 pKa = 10.64GLSVLHH458 pKa = 5.91PTIIDD463 pKa = 3.3NGAIYY468 pKa = 8.15PTDD471 pKa = 3.22ISSNILPLTPSFSIDD486 pKa = 3.39VNYY489 pKa = 10.76SDD491 pKa = 5.76YY492 pKa = 11.17NIHH495 pKa = 6.93PANIRR500 pKa = 11.84RR501 pKa = 11.84KK502 pKa = 8.5RR503 pKa = 11.84KK504 pKa = 9.36HH505 pKa = 6.09SSSLYY510 pKa = 9.47FF511 pKa = 3.99
Molecular weight: 55.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7B7D6|C7B7D6_9PAPI Putative E4 protein OS=Human papillomavirus 116 OX=915428 PE=4 SV=1
MM1 pKa = 6.92NQSDD5 pKa = 3.78LTEE8 pKa = 4.6RR9 pKa = 11.84LDD11 pKa = 4.06ALQTALMNIYY21 pKa = 10.27EE22 pKa = 4.51EE23 pKa = 5.02APTDD27 pKa = 3.64LPSQIRR33 pKa = 11.84HH34 pKa = 5.92FDD36 pKa = 3.78LLRR39 pKa = 11.84KK40 pKa = 9.8QSVLEE45 pKa = 3.91YY46 pKa = 9.14YY47 pKa = 9.93VRR49 pKa = 11.84KK50 pKa = 9.92EE51 pKa = 4.05GYY53 pKa = 6.85TQLGLYY59 pKa = 9.24HH60 pKa = 7.25IPTLKK65 pKa = 10.07VSEE68 pKa = 4.27YY69 pKa = 9.71HH70 pKa = 6.11AKK72 pKa = 10.11EE73 pKa = 4.22AIKK76 pKa = 9.97MGIVLRR82 pKa = 11.84SLQKK86 pKa = 10.46SPYY89 pKa = 10.13ADD91 pKa = 4.26EE92 pKa = 4.3EE93 pKa = 4.33WSLQDD98 pKa = 3.76VNADD102 pKa = 3.99FFNSPPRR109 pKa = 11.84NCFKK113 pKa = 10.91KK114 pKa = 10.11GGYY117 pKa = 9.23DD118 pKa = 3.31VEE120 pKa = 4.3VWFDD124 pKa = 3.96HH125 pKa = 6.75NPLNTFPYY133 pKa = 9.99TNWTWIYY140 pKa = 10.45YY141 pKa = 9.84QDD143 pKa = 5.8DD144 pKa = 3.78EE145 pKa = 4.96EE146 pKa = 4.48NWHH149 pKa = 6.0KK150 pKa = 11.03VQGEE154 pKa = 4.02TDD156 pKa = 3.79YY157 pKa = 11.87NGLFYY162 pKa = 10.93RR163 pKa = 11.84EE164 pKa = 3.67TDD166 pKa = 3.28GTVVYY171 pKa = 9.83FLLFEE176 pKa = 4.73SDD178 pKa = 2.89AARR181 pKa = 11.84YY182 pKa = 6.31GTKK185 pKa = 10.36NEE187 pKa = 3.55WTVNVKK193 pKa = 10.34NEE195 pKa = 4.31QISLPANSNGRR206 pKa = 11.84RR207 pKa = 11.84SSSGTPTHH215 pKa = 6.0STTNSVAEE223 pKa = 4.37PGPSRR228 pKa = 11.84STEE231 pKa = 3.86EE232 pKa = 3.6ADD234 pKa = 4.58GRR236 pKa = 11.84RR237 pKa = 11.84TPQAQTQSVGASKK250 pKa = 10.66KK251 pKa = 8.38PSSVGRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84RR261 pKa = 11.84KK262 pKa = 9.24QGKK265 pKa = 5.35QTPSKK270 pKa = 10.3RR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84TGGGSGDD280 pKa = 3.73EE281 pKa = 4.32ADD283 pKa = 3.96SGGISAEE290 pKa = 4.19EE291 pKa = 4.12VGSSHH296 pKa = 7.33RR297 pKa = 11.84SVARR301 pKa = 11.84SGLSRR306 pKa = 11.84LEE308 pKa = 3.96RR309 pKa = 11.84LQKK312 pKa = 9.91EE313 pKa = 3.89ARR315 pKa = 11.84DD316 pKa = 3.74PYY318 pKa = 10.26IILVRR323 pKa = 11.84GPQNTLKK330 pKa = 10.01CWRR333 pKa = 11.84YY334 pKa = 10.03RR335 pKa = 11.84IQTKK339 pKa = 10.85ANFPFLYY346 pKa = 10.13ISTVWKK352 pKa = 9.67WVTKK356 pKa = 10.4DD357 pKa = 3.4AVGHH361 pKa = 5.7EE362 pKa = 4.25GRR364 pKa = 11.84VLIAFLSKK372 pKa = 10.58EE373 pKa = 4.15SRR375 pKa = 11.84DD376 pKa = 3.48LFANSVHH383 pKa = 6.02FPKK386 pKa = 9.95NTTHH390 pKa = 7.24SYY392 pKa = 11.32GSLDD396 pKa = 3.38ALL398 pKa = 4.28
MM1 pKa = 6.92NQSDD5 pKa = 3.78LTEE8 pKa = 4.6RR9 pKa = 11.84LDD11 pKa = 4.06ALQTALMNIYY21 pKa = 10.27EE22 pKa = 4.51EE23 pKa = 5.02APTDD27 pKa = 3.64LPSQIRR33 pKa = 11.84HH34 pKa = 5.92FDD36 pKa = 3.78LLRR39 pKa = 11.84KK40 pKa = 9.8QSVLEE45 pKa = 3.91YY46 pKa = 9.14YY47 pKa = 9.93VRR49 pKa = 11.84KK50 pKa = 9.92EE51 pKa = 4.05GYY53 pKa = 6.85TQLGLYY59 pKa = 9.24HH60 pKa = 7.25IPTLKK65 pKa = 10.07VSEE68 pKa = 4.27YY69 pKa = 9.71HH70 pKa = 6.11AKK72 pKa = 10.11EE73 pKa = 4.22AIKK76 pKa = 9.97MGIVLRR82 pKa = 11.84SLQKK86 pKa = 10.46SPYY89 pKa = 10.13ADD91 pKa = 4.26EE92 pKa = 4.3EE93 pKa = 4.33WSLQDD98 pKa = 3.76VNADD102 pKa = 3.99FFNSPPRR109 pKa = 11.84NCFKK113 pKa = 10.91KK114 pKa = 10.11GGYY117 pKa = 9.23DD118 pKa = 3.31VEE120 pKa = 4.3VWFDD124 pKa = 3.96HH125 pKa = 6.75NPLNTFPYY133 pKa = 9.99TNWTWIYY140 pKa = 10.45YY141 pKa = 9.84QDD143 pKa = 5.8DD144 pKa = 3.78EE145 pKa = 4.96EE146 pKa = 4.48NWHH149 pKa = 6.0KK150 pKa = 11.03VQGEE154 pKa = 4.02TDD156 pKa = 3.79YY157 pKa = 11.87NGLFYY162 pKa = 10.93RR163 pKa = 11.84EE164 pKa = 3.67TDD166 pKa = 3.28GTVVYY171 pKa = 9.83FLLFEE176 pKa = 4.73SDD178 pKa = 2.89AARR181 pKa = 11.84YY182 pKa = 6.31GTKK185 pKa = 10.36NEE187 pKa = 3.55WTVNVKK193 pKa = 10.34NEE195 pKa = 4.31QISLPANSNGRR206 pKa = 11.84RR207 pKa = 11.84SSSGTPTHH215 pKa = 6.0STTNSVAEE223 pKa = 4.37PGPSRR228 pKa = 11.84STEE231 pKa = 3.86EE232 pKa = 3.6ADD234 pKa = 4.58GRR236 pKa = 11.84RR237 pKa = 11.84TPQAQTQSVGASKK250 pKa = 10.66KK251 pKa = 8.38PSSVGRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84RR261 pKa = 11.84KK262 pKa = 9.24QGKK265 pKa = 5.35QTPSKK270 pKa = 10.3RR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84TGGGSGDD280 pKa = 3.73EE281 pKa = 4.32ADD283 pKa = 3.96SGGISAEE290 pKa = 4.19EE291 pKa = 4.12VGSSHH296 pKa = 7.33RR297 pKa = 11.84SVARR301 pKa = 11.84SGLSRR306 pKa = 11.84LEE308 pKa = 3.96RR309 pKa = 11.84LQKK312 pKa = 9.91EE313 pKa = 3.89ARR315 pKa = 11.84DD316 pKa = 3.74PYY318 pKa = 10.26IILVRR323 pKa = 11.84GPQNTLKK330 pKa = 10.01CWRR333 pKa = 11.84YY334 pKa = 10.03RR335 pKa = 11.84IQTKK339 pKa = 10.85ANFPFLYY346 pKa = 10.13ISTVWKK352 pKa = 9.67WVTKK356 pKa = 10.4DD357 pKa = 3.4AVGHH361 pKa = 5.7EE362 pKa = 4.25GRR364 pKa = 11.84VLIAFLSKK372 pKa = 10.58EE373 pKa = 4.15SRR375 pKa = 11.84DD376 pKa = 3.48LFANSVHH383 pKa = 6.02FPKK386 pKa = 9.95NTTHH390 pKa = 7.24SYY392 pKa = 11.32GSLDD396 pKa = 3.38ALL398 pKa = 4.28
Molecular weight: 45.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2396 |
98 |
602 |
342.3 |
38.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.676 ± 0.27 | 2.337 ± 1.016 |
6.553 ± 0.294 | 6.386 ± 0.687 |
4.382 ± 0.539 | 5.676 ± 0.838 |
2.295 ± 0.194 | 4.967 ± 0.692 |
5.05 ± 0.761 | 9.641 ± 0.831 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.461 ± 0.27 | 5.092 ± 0.469 |
6.01 ± 1.006 | 4.007 ± 0.421 |
6.052 ± 0.807 | 7.095 ± 0.7 |
6.177 ± 0.617 | 6.344 ± 0.513 |
1.252 ± 0.321 | 3.548 ± 0.391 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
