
Suffolk virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Jingchuvirales; Chuviridae; Mivirus; Mivirus suffolkense
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

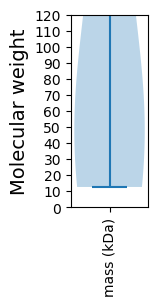
Protein with the lowest isoelectric point:
>tr|A0A0A1G6U7|A0A0A1G6U7_9VIRU Large structural protein OS=Suffolk virus OX=1577137 PE=4 SV=1
MM1 pKa = 6.73STSGRR6 pKa = 11.84SLSSCMLWVILCVLGGGQQAEE27 pKa = 4.3GLVGFDD33 pKa = 3.64CTGQAVNVTAVSLSWIQKK51 pKa = 9.34CPVAEE56 pKa = 4.55GAQGTIVEE64 pKa = 5.42FIQLVQEE71 pKa = 4.4RR72 pKa = 11.84NVEE75 pKa = 4.05AVRR78 pKa = 11.84VKK80 pKa = 10.36ACLVEE85 pKa = 4.55RR86 pKa = 11.84SYY88 pKa = 11.76LLQHH92 pKa = 6.89CGMHH96 pKa = 5.76SHH98 pKa = 7.01SSMTFNGLVTSEE110 pKa = 3.9ILRR113 pKa = 11.84VPKK116 pKa = 9.66EE117 pKa = 4.02ACDD120 pKa = 3.63ALHH123 pKa = 7.1LYY125 pKa = 10.9GSFHH129 pKa = 6.29TSGGKK134 pKa = 9.64VLTGFAANTTTVVPVVEE151 pKa = 4.66MGVIDD156 pKa = 5.4AGSATCKK163 pKa = 10.46GATFTLNGATYY174 pKa = 9.54TDD176 pKa = 3.55VVMQSSYY183 pKa = 11.21KK184 pKa = 8.77VTLRR188 pKa = 11.84TEE190 pKa = 4.39RR191 pKa = 11.84AQLDD195 pKa = 3.99VATSTVRR202 pKa = 11.84LSSGYY207 pKa = 8.12SHH209 pKa = 7.98PFQAHH214 pKa = 5.83SGFDD218 pKa = 3.52PEE220 pKa = 5.68LGQTYY225 pKa = 9.03WSSEE229 pKa = 3.91GVGVKK234 pKa = 10.29CSPTSYY240 pKa = 9.74IVVYY244 pKa = 10.37EE245 pKa = 4.18GDD247 pKa = 2.99ATIYY251 pKa = 10.51KK252 pKa = 9.84NRR254 pKa = 11.84EE255 pKa = 3.75GQRR258 pKa = 11.84TLIVNTTTQVMAVGLRR274 pKa = 11.84DD275 pKa = 3.72EE276 pKa = 4.56TTLCHH281 pKa = 6.72QEE283 pKa = 3.89ATEE286 pKa = 4.15TDD288 pKa = 3.42HH289 pKa = 7.34PRR291 pKa = 11.84LFMVRR296 pKa = 11.84PLRR299 pKa = 11.84AGGPRR304 pKa = 11.84FYY306 pKa = 10.45FQKK309 pKa = 11.09GSMDD313 pKa = 3.7PKK315 pKa = 10.69EE316 pKa = 4.19VDD318 pKa = 2.99LFLYY322 pKa = 9.58TNSKK326 pKa = 9.61LVYY329 pKa = 9.88VEE331 pKa = 3.64QHH333 pKa = 6.51LARR336 pKa = 11.84EE337 pKa = 4.24LTSVYY342 pKa = 11.06LHH344 pKa = 6.3FQKK347 pKa = 10.64QMCDD351 pKa = 3.11VNHH354 pKa = 7.18RR355 pKa = 11.84LLTHH359 pKa = 6.56LTTLAMVAPEE369 pKa = 4.05EE370 pKa = 4.23FAWTYY375 pKa = 6.94TQQPGITAVLRR386 pKa = 11.84GEE388 pKa = 4.05VVYY391 pKa = 9.37MVKK394 pKa = 10.35CAPVSVDD401 pKa = 3.12YY402 pKa = 10.46RR403 pKa = 11.84ASSACFQEE411 pKa = 5.13IPVTYY416 pKa = 10.51NGTQVFLKK424 pKa = 9.5PRR426 pKa = 11.84SRR428 pKa = 11.84IITTYY433 pKa = 9.14GTEE436 pKa = 4.1IDD438 pKa = 4.71CSPLAPPLFFLNGNWVSFAPHH459 pKa = 6.56PTHH462 pKa = 6.66VLPPQVLNASFQAEE476 pKa = 4.09WAYY479 pKa = 9.78TSPPHH484 pKa = 6.94LISAGLYY491 pKa = 9.28SQSLLEE497 pKa = 4.3KK498 pKa = 10.09YY499 pKa = 9.49QRR501 pKa = 11.84QLMFPIEE508 pKa = 3.96RR509 pKa = 11.84TAIEE513 pKa = 4.16TTMAASMAGLNVDD526 pKa = 3.81HH527 pKa = 6.98QGLDD531 pKa = 2.98ASGLLRR537 pKa = 11.84QSGLDD542 pKa = 3.39KK543 pKa = 10.98LQTSFMEE550 pKa = 5.79RR551 pKa = 11.84IYY553 pKa = 10.81GWWWTFSVNLAGVMGVIFVITAIKK577 pKa = 10.47AIGNVVLNATFLYY590 pKa = 9.11KK591 pKa = 10.49TFGCGFRR598 pKa = 11.84MFAMFWGTLAKK609 pKa = 10.46YY610 pKa = 10.5LLIKK614 pKa = 10.77VPTNPSRR621 pKa = 11.84DD622 pKa = 3.78DD623 pKa = 3.69VEE625 pKa = 6.36AGTPSPPTSSPTPCSSGACNEE646 pKa = 3.98EE647 pKa = 3.77ATILISPPSSHH658 pKa = 7.57FYY660 pKa = 10.21PSVSYY665 pKa = 10.57RR666 pKa = 11.84PPGATAPIII675 pKa = 3.95
MM1 pKa = 6.73STSGRR6 pKa = 11.84SLSSCMLWVILCVLGGGQQAEE27 pKa = 4.3GLVGFDD33 pKa = 3.64CTGQAVNVTAVSLSWIQKK51 pKa = 9.34CPVAEE56 pKa = 4.55GAQGTIVEE64 pKa = 5.42FIQLVQEE71 pKa = 4.4RR72 pKa = 11.84NVEE75 pKa = 4.05AVRR78 pKa = 11.84VKK80 pKa = 10.36ACLVEE85 pKa = 4.55RR86 pKa = 11.84SYY88 pKa = 11.76LLQHH92 pKa = 6.89CGMHH96 pKa = 5.76SHH98 pKa = 7.01SSMTFNGLVTSEE110 pKa = 3.9ILRR113 pKa = 11.84VPKK116 pKa = 9.66EE117 pKa = 4.02ACDD120 pKa = 3.63ALHH123 pKa = 7.1LYY125 pKa = 10.9GSFHH129 pKa = 6.29TSGGKK134 pKa = 9.64VLTGFAANTTTVVPVVEE151 pKa = 4.66MGVIDD156 pKa = 5.4AGSATCKK163 pKa = 10.46GATFTLNGATYY174 pKa = 9.54TDD176 pKa = 3.55VVMQSSYY183 pKa = 11.21KK184 pKa = 8.77VTLRR188 pKa = 11.84TEE190 pKa = 4.39RR191 pKa = 11.84AQLDD195 pKa = 3.99VATSTVRR202 pKa = 11.84LSSGYY207 pKa = 8.12SHH209 pKa = 7.98PFQAHH214 pKa = 5.83SGFDD218 pKa = 3.52PEE220 pKa = 5.68LGQTYY225 pKa = 9.03WSSEE229 pKa = 3.91GVGVKK234 pKa = 10.29CSPTSYY240 pKa = 9.74IVVYY244 pKa = 10.37EE245 pKa = 4.18GDD247 pKa = 2.99ATIYY251 pKa = 10.51KK252 pKa = 9.84NRR254 pKa = 11.84EE255 pKa = 3.75GQRR258 pKa = 11.84TLIVNTTTQVMAVGLRR274 pKa = 11.84DD275 pKa = 3.72EE276 pKa = 4.56TTLCHH281 pKa = 6.72QEE283 pKa = 3.89ATEE286 pKa = 4.15TDD288 pKa = 3.42HH289 pKa = 7.34PRR291 pKa = 11.84LFMVRR296 pKa = 11.84PLRR299 pKa = 11.84AGGPRR304 pKa = 11.84FYY306 pKa = 10.45FQKK309 pKa = 11.09GSMDD313 pKa = 3.7PKK315 pKa = 10.69EE316 pKa = 4.19VDD318 pKa = 2.99LFLYY322 pKa = 9.58TNSKK326 pKa = 9.61LVYY329 pKa = 9.88VEE331 pKa = 3.64QHH333 pKa = 6.51LARR336 pKa = 11.84EE337 pKa = 4.24LTSVYY342 pKa = 11.06LHH344 pKa = 6.3FQKK347 pKa = 10.64QMCDD351 pKa = 3.11VNHH354 pKa = 7.18RR355 pKa = 11.84LLTHH359 pKa = 6.56LTTLAMVAPEE369 pKa = 4.05EE370 pKa = 4.23FAWTYY375 pKa = 6.94TQQPGITAVLRR386 pKa = 11.84GEE388 pKa = 4.05VVYY391 pKa = 9.37MVKK394 pKa = 10.35CAPVSVDD401 pKa = 3.12YY402 pKa = 10.46RR403 pKa = 11.84ASSACFQEE411 pKa = 5.13IPVTYY416 pKa = 10.51NGTQVFLKK424 pKa = 9.5PRR426 pKa = 11.84SRR428 pKa = 11.84IITTYY433 pKa = 9.14GTEE436 pKa = 4.1IDD438 pKa = 4.71CSPLAPPLFFLNGNWVSFAPHH459 pKa = 6.56PTHH462 pKa = 6.66VLPPQVLNASFQAEE476 pKa = 4.09WAYY479 pKa = 9.78TSPPHH484 pKa = 6.94LISAGLYY491 pKa = 9.28SQSLLEE497 pKa = 4.3KK498 pKa = 10.09YY499 pKa = 9.49QRR501 pKa = 11.84QLMFPIEE508 pKa = 3.96RR509 pKa = 11.84TAIEE513 pKa = 4.16TTMAASMAGLNVDD526 pKa = 3.81HH527 pKa = 6.98QGLDD531 pKa = 2.98ASGLLRR537 pKa = 11.84QSGLDD542 pKa = 3.39KK543 pKa = 10.98LQTSFMEE550 pKa = 5.79RR551 pKa = 11.84IYY553 pKa = 10.81GWWWTFSVNLAGVMGVIFVITAIKK577 pKa = 10.47AIGNVVLNATFLYY590 pKa = 9.11KK591 pKa = 10.49TFGCGFRR598 pKa = 11.84MFAMFWGTLAKK609 pKa = 10.46YY610 pKa = 10.5LLIKK614 pKa = 10.77VPTNPSRR621 pKa = 11.84DD622 pKa = 3.78DD623 pKa = 3.69VEE625 pKa = 6.36AGTPSPPTSSPTPCSSGACNEE646 pKa = 3.98EE647 pKa = 3.77ATILISPPSSHH658 pKa = 7.57FYY660 pKa = 10.21PSVSYY665 pKa = 10.57RR666 pKa = 11.84PPGATAPIII675 pKa = 3.95
Molecular weight: 73.95 kDa
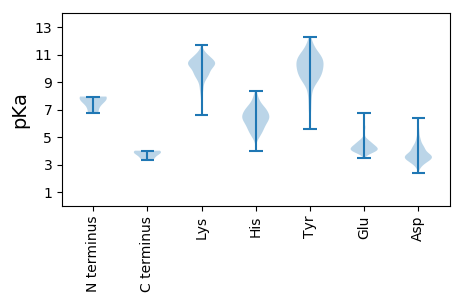
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1G690|A0A0A1G690_9VIRU Putative glycoprotein OS=Suffolk virus OX=1577137 PE=4 SV=1
MM1 pKa = 7.94GDD3 pKa = 3.65PVVQPVVVEE12 pKa = 4.52AGARR16 pKa = 11.84AQLQAAPPGFWNPRR30 pKa = 11.84DD31 pKa = 3.47TEE33 pKa = 4.19YY34 pKa = 11.0QRR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.15AFGGIVATDD47 pKa = 3.38QLTGANCDD55 pKa = 4.04FLAGTSDD62 pKa = 3.58DD63 pKa = 5.02AIFPRR68 pKa = 11.84ITIMARR74 pKa = 11.84GEE76 pKa = 4.22AYY78 pKa = 10.54LKK80 pKa = 10.71AITDD84 pKa = 3.62IEE86 pKa = 4.45KK87 pKa = 9.45ATLTIAIANAIAPNMTALALPPAEE111 pKa = 3.9ASARR115 pKa = 11.84GRR117 pKa = 11.84TQVVEE122 pKa = 4.85FLRR125 pKa = 11.84QEE127 pKa = 3.72VGKK130 pKa = 9.86NVKK133 pKa = 9.09DD134 pKa = 3.51LCVITPQLWASARR147 pKa = 11.84AAEE150 pKa = 4.9GPLHH154 pKa = 7.03RR155 pKa = 11.84IPWLCDD161 pKa = 3.72FNGLPTLVRR170 pKa = 11.84PADD173 pKa = 3.67GTDD176 pKa = 3.24TVGHH180 pKa = 6.1YY181 pKa = 10.96NSILAFLLTYY191 pKa = 6.63PTPYY195 pKa = 9.57ISNIGTRR202 pKa = 11.84LYY204 pKa = 10.84AILYY208 pKa = 9.81VSLAKK213 pKa = 10.39QGTISRR219 pKa = 11.84GKK221 pKa = 10.24LEE223 pKa = 5.8KK224 pKa = 10.23INQDD228 pKa = 3.36LAVNMGFEE236 pKa = 4.55VNMTEE241 pKa = 4.22EE242 pKa = 4.74VITYY246 pKa = 7.6TYY248 pKa = 10.0QQIGTKK254 pKa = 9.5IPHH257 pKa = 6.32NALEE261 pKa = 4.23IMFNTMRR268 pKa = 11.84DD269 pKa = 3.2HH270 pKa = 5.94MQNLSLRR277 pKa = 11.84MQVTLQQAAGTGLTGLQIIKK297 pKa = 10.16RR298 pKa = 11.84AILEE302 pKa = 4.27HH303 pKa = 6.98PAFPWGKK310 pKa = 9.12LAQMLPAEE318 pKa = 4.6GPKK321 pKa = 10.4VIAAFEE327 pKa = 4.14AVGNDD332 pKa = 3.43PFYY335 pKa = 11.54GFLPDD340 pKa = 4.73LGPAKK345 pKa = 9.21STNYY349 pKa = 9.86ARR351 pKa = 11.84YY352 pKa = 8.73VWVCQRR358 pKa = 11.84LLRR361 pKa = 11.84KK362 pKa = 9.81FNAEE366 pKa = 3.92DD367 pKa = 3.47EE368 pKa = 4.5RR369 pKa = 11.84TLRR372 pKa = 11.84NYY374 pKa = 10.44KK375 pKa = 10.39GGVRR379 pKa = 11.84GIPNQPLFEE388 pKa = 4.33EE389 pKa = 5.78LIDD392 pKa = 4.2SYY394 pKa = 11.72SPPAPEE400 pKa = 4.38AAVSADD406 pKa = 3.73FVALGVNITALARR419 pKa = 11.84SCRR422 pKa = 11.84ALQSS426 pKa = 3.36
MM1 pKa = 7.94GDD3 pKa = 3.65PVVQPVVVEE12 pKa = 4.52AGARR16 pKa = 11.84AQLQAAPPGFWNPRR30 pKa = 11.84DD31 pKa = 3.47TEE33 pKa = 4.19YY34 pKa = 11.0QRR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.15AFGGIVATDD47 pKa = 3.38QLTGANCDD55 pKa = 4.04FLAGTSDD62 pKa = 3.58DD63 pKa = 5.02AIFPRR68 pKa = 11.84ITIMARR74 pKa = 11.84GEE76 pKa = 4.22AYY78 pKa = 10.54LKK80 pKa = 10.71AITDD84 pKa = 3.62IEE86 pKa = 4.45KK87 pKa = 9.45ATLTIAIANAIAPNMTALALPPAEE111 pKa = 3.9ASARR115 pKa = 11.84GRR117 pKa = 11.84TQVVEE122 pKa = 4.85FLRR125 pKa = 11.84QEE127 pKa = 3.72VGKK130 pKa = 9.86NVKK133 pKa = 9.09DD134 pKa = 3.51LCVITPQLWASARR147 pKa = 11.84AAEE150 pKa = 4.9GPLHH154 pKa = 7.03RR155 pKa = 11.84IPWLCDD161 pKa = 3.72FNGLPTLVRR170 pKa = 11.84PADD173 pKa = 3.67GTDD176 pKa = 3.24TVGHH180 pKa = 6.1YY181 pKa = 10.96NSILAFLLTYY191 pKa = 6.63PTPYY195 pKa = 9.57ISNIGTRR202 pKa = 11.84LYY204 pKa = 10.84AILYY208 pKa = 9.81VSLAKK213 pKa = 10.39QGTISRR219 pKa = 11.84GKK221 pKa = 10.24LEE223 pKa = 5.8KK224 pKa = 10.23INQDD228 pKa = 3.36LAVNMGFEE236 pKa = 4.55VNMTEE241 pKa = 4.22EE242 pKa = 4.74VITYY246 pKa = 7.6TYY248 pKa = 10.0QQIGTKK254 pKa = 9.5IPHH257 pKa = 6.32NALEE261 pKa = 4.23IMFNTMRR268 pKa = 11.84DD269 pKa = 3.2HH270 pKa = 5.94MQNLSLRR277 pKa = 11.84MQVTLQQAAGTGLTGLQIIKK297 pKa = 10.16RR298 pKa = 11.84AILEE302 pKa = 4.27HH303 pKa = 6.98PAFPWGKK310 pKa = 9.12LAQMLPAEE318 pKa = 4.6GPKK321 pKa = 10.4VIAAFEE327 pKa = 4.14AVGNDD332 pKa = 3.43PFYY335 pKa = 11.54GFLPDD340 pKa = 4.73LGPAKK345 pKa = 9.21STNYY349 pKa = 9.86ARR351 pKa = 11.84YY352 pKa = 8.73VWVCQRR358 pKa = 11.84LLRR361 pKa = 11.84KK362 pKa = 9.81FNAEE366 pKa = 3.92DD367 pKa = 3.47EE368 pKa = 4.5RR369 pKa = 11.84TLRR372 pKa = 11.84NYY374 pKa = 10.44KK375 pKa = 10.39GGVRR379 pKa = 11.84GIPNQPLFEE388 pKa = 4.33EE389 pKa = 5.78LIDD392 pKa = 4.2SYY394 pKa = 11.72SPPAPEE400 pKa = 4.38AAVSADD406 pKa = 3.73FVALGVNITALARR419 pKa = 11.84SCRR422 pKa = 11.84ALQSS426 pKa = 3.36
Molecular weight: 46.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3382 |
113 |
2168 |
845.5 |
94.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.658 ± 1.353 | 2.07 ± 0.356 |
4.879 ± 0.824 | 5.707 ± 0.441 |
3.578 ± 0.471 | 6.062 ± 0.668 |
2.543 ± 0.402 | 4.967 ± 0.522 |
4.021 ± 0.469 | 10.408 ± 1.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.602 ± 0.151 | 3.459 ± 0.474 |
5.795 ± 1.001 | 3.637 ± 0.582 |
6.091 ± 0.729 | 5.943 ± 0.93 |
7.126 ± 0.739 | 7.895 ± 0.584 |
1.626 ± 0.283 | 3.933 ± 0.277 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
