
Acidobacteria bacterium SCGC AG-212-P17
Taxonomy: cellular organisms; Bacteria; Acidobacteria; unclassified Acidobacteria
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
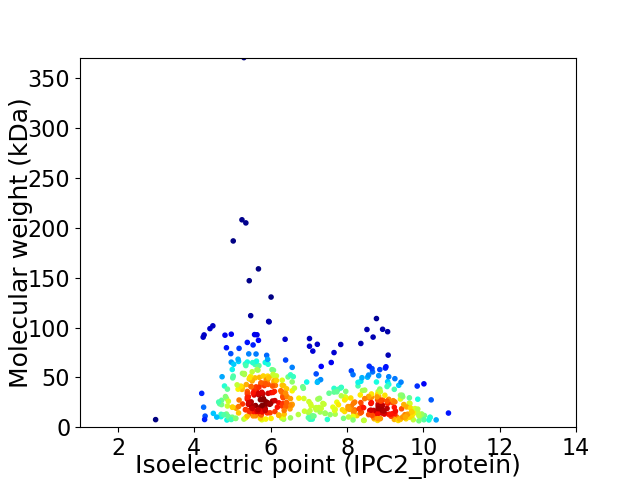
Virtual 2D-PAGE plot for 481 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A177RFP3|A0A177RFP3_9BACT Lon protease OS=Acidobacteria bacterium SCGC AG-212-P17 OX=1803033 GN=lon PE=2 SV=1
MM1 pKa = 7.43FFLLVASGILFLPKK15 pKa = 10.14FSLADD20 pKa = 3.42GVIIQTPAQGHH31 pKa = 4.91TVSITHH37 pKa = 6.06TVSSFDD43 pKa = 3.37IGTAAANVTIANTGMWISFNPGISQANFCGRR74 pKa = 11.84SDD76 pKa = 4.77FGVLDD81 pKa = 4.03INLHH85 pKa = 6.73DD86 pKa = 4.0NTTGQDD92 pKa = 3.24AYY94 pKa = 11.11SGQLAFDD101 pKa = 3.32QGVYY105 pKa = 9.16GTTGLSHH112 pKa = 6.72IHH114 pKa = 7.11DD115 pKa = 3.97VTAGDD120 pKa = 5.69DD121 pKa = 3.37IRR123 pKa = 11.84IDD125 pKa = 4.07LNCDD129 pKa = 2.99PSSATTTITAGITDD143 pKa = 4.05IPSVYY148 pKa = 7.92WWNGSGMTQPQYY160 pKa = 11.61DD161 pKa = 3.45AMIAYY166 pKa = 7.32MTSQLTVSSLHH177 pKa = 5.75QYY179 pKa = 10.59KK180 pKa = 10.4SDD182 pKa = 3.06ASTNIAEE189 pKa = 4.86GGLTTEE195 pKa = 4.04NSLVIKK201 pKa = 8.91GIPTVASGTDD211 pKa = 3.26QLRR214 pKa = 11.84LEE216 pKa = 5.21LEE218 pKa = 4.25VQPFATAFTGTATATSAATSSGNVVSLTIPSLANNHH254 pKa = 4.68YY255 pKa = 9.04HH256 pKa = 4.99WRR258 pKa = 11.84ARR260 pKa = 11.84VFDD263 pKa = 3.8TTTNATSSWEE273 pKa = 3.98EE274 pKa = 3.86FGTSGNADD282 pKa = 3.54FTIHH286 pKa = 7.06AGGNTADD293 pKa = 3.81LYY295 pKa = 11.57YY296 pKa = 10.15PVGYY300 pKa = 10.12QWISKK305 pKa = 8.36NAQYY309 pKa = 10.69CYY311 pKa = 11.0DD312 pKa = 3.96HH313 pKa = 7.17FSTSSPACSGVTSATSSDD331 pKa = 3.07NVYY334 pKa = 10.15WGGLVSDD341 pKa = 3.93NTAISFSGYY350 pKa = 10.42VSMLLYY356 pKa = 10.5DD357 pKa = 4.37ASSTSQCASGWWGHH371 pKa = 5.38TQDD374 pKa = 4.75IVDD377 pKa = 4.05ASGTLTDD384 pKa = 3.89VFDD387 pKa = 4.7GSVAAVGGVICHH399 pKa = 6.18NAGGAKK405 pKa = 9.5TISGPLTSRR414 pKa = 11.84RR415 pKa = 11.84YY416 pKa = 8.96VVVWQDD422 pKa = 3.55PAHH425 pKa = 6.36KK426 pKa = 10.29CADD429 pKa = 3.64EE430 pKa = 4.66SLSQCLADD438 pKa = 4.79HH439 pKa = 6.78GGDD442 pKa = 3.43SDD444 pKa = 4.96FSLTTLSFDD453 pKa = 3.91PVSPPPDD460 pKa = 3.43NARR463 pKa = 11.84SIYY466 pKa = 10.59LDD468 pKa = 3.49GSSALGYY475 pKa = 9.94SAANVAFTATDD486 pKa = 3.64PFTIEE491 pKa = 3.13LWYY494 pKa = 10.66RR495 pKa = 11.84SIAPSSTVMKK505 pKa = 10.67FIDD508 pKa = 3.56TRR510 pKa = 11.84SGQGKK515 pKa = 9.06GFFLEE520 pKa = 4.02KK521 pKa = 10.05HH522 pKa = 6.12ANDD525 pKa = 4.8GIEE528 pKa = 4.58FFVACDD534 pKa = 3.14SGEE537 pKa = 4.2VLFHH541 pKa = 6.67GAGALADD548 pKa = 4.7DD549 pKa = 4.68LTLNPAFDD557 pKa = 3.57KK558 pKa = 11.02HH559 pKa = 6.1GVWHH563 pKa = 6.55HH564 pKa = 5.05VAITKK569 pKa = 10.49DD570 pKa = 3.52SGTSWTAFKK579 pKa = 10.74LYY581 pKa = 10.41FDD583 pKa = 5.93GISQSISMDD592 pKa = 3.12TSNTLSGSCFNSTSSDD608 pKa = 3.88AIWFGQNQNPSSTPTFFTGNLDD630 pKa = 3.58EE631 pKa = 4.11VRR633 pKa = 11.84IWNVEE638 pKa = 3.8KK639 pKa = 11.01SVDD642 pKa = 4.02DD643 pKa = 4.09IKK645 pKa = 11.53AAMATEE651 pKa = 4.31ATSTAGLLALWQFNNTSTNIVTSDD675 pKa = 3.34VASGQDD681 pKa = 3.53GMPVFATSTPFGHH694 pKa = 6.78FLIDD698 pKa = 3.2QTVYY702 pKa = 8.35PTSVRR707 pKa = 11.84DD708 pKa = 3.81GQLLYY713 pKa = 10.72YY714 pKa = 10.7SSGTSYY720 pKa = 11.0GSEE723 pKa = 3.9MSDD726 pKa = 4.99AASTWNALGDD736 pKa = 3.83LVIASTTSALSANVVVSDD754 pKa = 3.98VNSNSSDD761 pKa = 3.96YY762 pKa = 10.92AWEE765 pKa = 4.59GAWTPYY771 pKa = 7.86PTSSLPSLRR780 pKa = 11.84FNHH783 pKa = 6.6YY784 pKa = 8.22YY785 pKa = 9.47TDD787 pKa = 3.33SRR789 pKa = 11.84SYY791 pKa = 11.45DD792 pKa = 3.69EE793 pKa = 4.56IMHH796 pKa = 5.96VATHH800 pKa = 6.06EE801 pKa = 4.34LGHH804 pKa = 7.1AIGLDD809 pKa = 3.49HH810 pKa = 7.12SFLGNIMNYY819 pKa = 9.86YY820 pKa = 8.76VTAQTFLGGQDD831 pKa = 2.99QKK833 pKa = 11.89DD834 pKa = 3.54YY835 pKa = 11.54FFINDD840 pKa = 3.15EE841 pKa = 4.32GLWGNN846 pKa = 3.97
MM1 pKa = 7.43FFLLVASGILFLPKK15 pKa = 10.14FSLADD20 pKa = 3.42GVIIQTPAQGHH31 pKa = 4.91TVSITHH37 pKa = 6.06TVSSFDD43 pKa = 3.37IGTAAANVTIANTGMWISFNPGISQANFCGRR74 pKa = 11.84SDD76 pKa = 4.77FGVLDD81 pKa = 4.03INLHH85 pKa = 6.73DD86 pKa = 4.0NTTGQDD92 pKa = 3.24AYY94 pKa = 11.11SGQLAFDD101 pKa = 3.32QGVYY105 pKa = 9.16GTTGLSHH112 pKa = 6.72IHH114 pKa = 7.11DD115 pKa = 3.97VTAGDD120 pKa = 5.69DD121 pKa = 3.37IRR123 pKa = 11.84IDD125 pKa = 4.07LNCDD129 pKa = 2.99PSSATTTITAGITDD143 pKa = 4.05IPSVYY148 pKa = 7.92WWNGSGMTQPQYY160 pKa = 11.61DD161 pKa = 3.45AMIAYY166 pKa = 7.32MTSQLTVSSLHH177 pKa = 5.75QYY179 pKa = 10.59KK180 pKa = 10.4SDD182 pKa = 3.06ASTNIAEE189 pKa = 4.86GGLTTEE195 pKa = 4.04NSLVIKK201 pKa = 8.91GIPTVASGTDD211 pKa = 3.26QLRR214 pKa = 11.84LEE216 pKa = 5.21LEE218 pKa = 4.25VQPFATAFTGTATATSAATSSGNVVSLTIPSLANNHH254 pKa = 4.68YY255 pKa = 9.04HH256 pKa = 4.99WRR258 pKa = 11.84ARR260 pKa = 11.84VFDD263 pKa = 3.8TTTNATSSWEE273 pKa = 3.98EE274 pKa = 3.86FGTSGNADD282 pKa = 3.54FTIHH286 pKa = 7.06AGGNTADD293 pKa = 3.81LYY295 pKa = 11.57YY296 pKa = 10.15PVGYY300 pKa = 10.12QWISKK305 pKa = 8.36NAQYY309 pKa = 10.69CYY311 pKa = 11.0DD312 pKa = 3.96HH313 pKa = 7.17FSTSSPACSGVTSATSSDD331 pKa = 3.07NVYY334 pKa = 10.15WGGLVSDD341 pKa = 3.93NTAISFSGYY350 pKa = 10.42VSMLLYY356 pKa = 10.5DD357 pKa = 4.37ASSTSQCASGWWGHH371 pKa = 5.38TQDD374 pKa = 4.75IVDD377 pKa = 4.05ASGTLTDD384 pKa = 3.89VFDD387 pKa = 4.7GSVAAVGGVICHH399 pKa = 6.18NAGGAKK405 pKa = 9.5TISGPLTSRR414 pKa = 11.84RR415 pKa = 11.84YY416 pKa = 8.96VVVWQDD422 pKa = 3.55PAHH425 pKa = 6.36KK426 pKa = 10.29CADD429 pKa = 3.64EE430 pKa = 4.66SLSQCLADD438 pKa = 4.79HH439 pKa = 6.78GGDD442 pKa = 3.43SDD444 pKa = 4.96FSLTTLSFDD453 pKa = 3.91PVSPPPDD460 pKa = 3.43NARR463 pKa = 11.84SIYY466 pKa = 10.59LDD468 pKa = 3.49GSSALGYY475 pKa = 9.94SAANVAFTATDD486 pKa = 3.64PFTIEE491 pKa = 3.13LWYY494 pKa = 10.66RR495 pKa = 11.84SIAPSSTVMKK505 pKa = 10.67FIDD508 pKa = 3.56TRR510 pKa = 11.84SGQGKK515 pKa = 9.06GFFLEE520 pKa = 4.02KK521 pKa = 10.05HH522 pKa = 6.12ANDD525 pKa = 4.8GIEE528 pKa = 4.58FFVACDD534 pKa = 3.14SGEE537 pKa = 4.2VLFHH541 pKa = 6.67GAGALADD548 pKa = 4.7DD549 pKa = 4.68LTLNPAFDD557 pKa = 3.57KK558 pKa = 11.02HH559 pKa = 6.1GVWHH563 pKa = 6.55HH564 pKa = 5.05VAITKK569 pKa = 10.49DD570 pKa = 3.52SGTSWTAFKK579 pKa = 10.74LYY581 pKa = 10.41FDD583 pKa = 5.93GISQSISMDD592 pKa = 3.12TSNTLSGSCFNSTSSDD608 pKa = 3.88AIWFGQNQNPSSTPTFFTGNLDD630 pKa = 3.58EE631 pKa = 4.11VRR633 pKa = 11.84IWNVEE638 pKa = 3.8KK639 pKa = 11.01SVDD642 pKa = 4.02DD643 pKa = 4.09IKK645 pKa = 11.53AAMATEE651 pKa = 4.31ATSTAGLLALWQFNNTSTNIVTSDD675 pKa = 3.34VASGQDD681 pKa = 3.53GMPVFATSTPFGHH694 pKa = 6.78FLIDD698 pKa = 3.2QTVYY702 pKa = 8.35PTSVRR707 pKa = 11.84DD708 pKa = 3.81GQLLYY713 pKa = 10.72YY714 pKa = 10.7SSGTSYY720 pKa = 11.0GSEE723 pKa = 3.9MSDD726 pKa = 4.99AASTWNALGDD736 pKa = 3.83LVIASTTSALSANVVVSDD754 pKa = 3.98VNSNSSDD761 pKa = 3.96YY762 pKa = 10.92AWEE765 pKa = 4.59GAWTPYY771 pKa = 7.86PTSSLPSLRR780 pKa = 11.84FNHH783 pKa = 6.6YY784 pKa = 8.22YY785 pKa = 9.47TDD787 pKa = 3.33SRR789 pKa = 11.84SYY791 pKa = 11.45DD792 pKa = 3.69EE793 pKa = 4.56IMHH796 pKa = 5.96VATHH800 pKa = 6.06EE801 pKa = 4.34LGHH804 pKa = 7.1AIGLDD809 pKa = 3.49HH810 pKa = 7.12SFLGNIMNYY819 pKa = 9.86YY820 pKa = 8.76VTAQTFLGGQDD831 pKa = 2.99QKK833 pKa = 11.89DD834 pKa = 3.54YY835 pKa = 11.54FFINDD840 pKa = 3.15EE841 pKa = 4.32GLWGNN846 pKa = 3.97
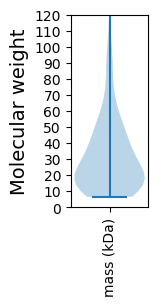
Molecular weight: 90.37 kDa
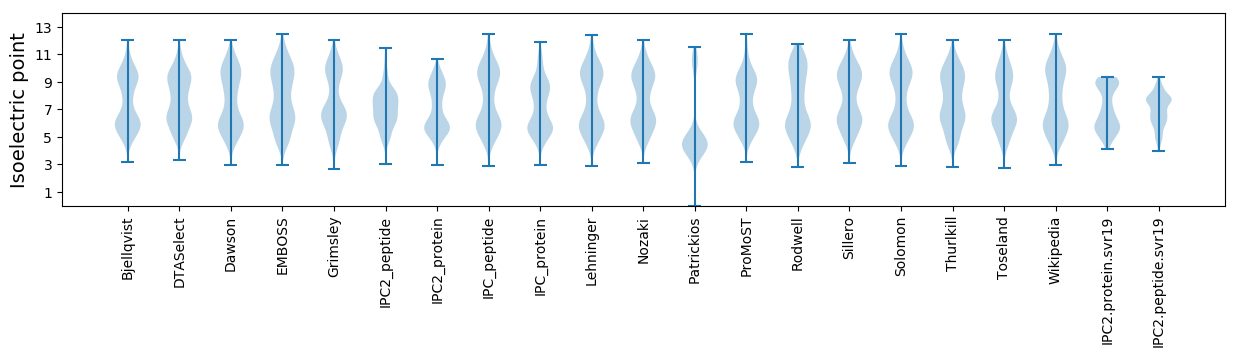
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177RCU4|A0A177RCU4_9BACT Uncharacterized protein OS=Acidobacteria bacterium SCGC AG-212-P17 OX=1803033 GN=AYO50_01475 PE=4 SV=1
MM1 pKa = 7.39NNATCCMMTTSRR13 pKa = 11.84VHH15 pKa = 5.68TSAGKK20 pKa = 10.0AADD23 pKa = 3.6GKK25 pKa = 10.65ARR27 pKa = 11.84PSSAGRR33 pKa = 11.84RR34 pKa = 11.84GFAVGKK40 pKa = 9.92FSLPMLILALLPKK53 pKa = 10.54CPACFAAYY61 pKa = 9.65LALGTGISVSVTVASLFRR79 pKa = 11.84TLLIGICVTSLVWIFASAFRR99 pKa = 11.84SALMNRR105 pKa = 11.84LRR107 pKa = 11.84SDD109 pKa = 2.95SRR111 pKa = 11.84LYY113 pKa = 10.92
MM1 pKa = 7.39NNATCCMMTTSRR13 pKa = 11.84VHH15 pKa = 5.68TSAGKK20 pKa = 10.0AADD23 pKa = 3.6GKK25 pKa = 10.65ARR27 pKa = 11.84PSSAGRR33 pKa = 11.84RR34 pKa = 11.84GFAVGKK40 pKa = 9.92FSLPMLILALLPKK53 pKa = 10.54CPACFAAYY61 pKa = 9.65LALGTGISVSVTVASLFRR79 pKa = 11.84TLLIGICVTSLVWIFASAFRR99 pKa = 11.84SALMNRR105 pKa = 11.84LRR107 pKa = 11.84SDD109 pKa = 2.95SRR111 pKa = 11.84LYY113 pKa = 10.92
Molecular weight: 11.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
150889 |
59 |
3347 |
313.7 |
34.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.245 ± 0.137 | 0.956 ± 0.039 |
5.02 ± 0.07 | 5.664 ± 0.138 |
4.123 ± 0.069 | 7.538 ± 0.099 |
2.21 ± 0.05 | 5.242 ± 0.091 |
4.506 ± 0.111 | 9.846 ± 0.133 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.268 ± 0.056 | 3.585 ± 0.081 |
5.287 ± 0.109 | 4.124 ± 0.077 |
5.928 ± 0.097 | 6.512 ± 0.106 |
5.724 ± 0.113 | 7.269 ± 0.085 |
1.35 ± 0.038 | 2.602 ± 0.062 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
