
Levilinea saccharolytica
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Anaerolineae; Anaerolineales; Anaerolineaceae; Levilinea
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
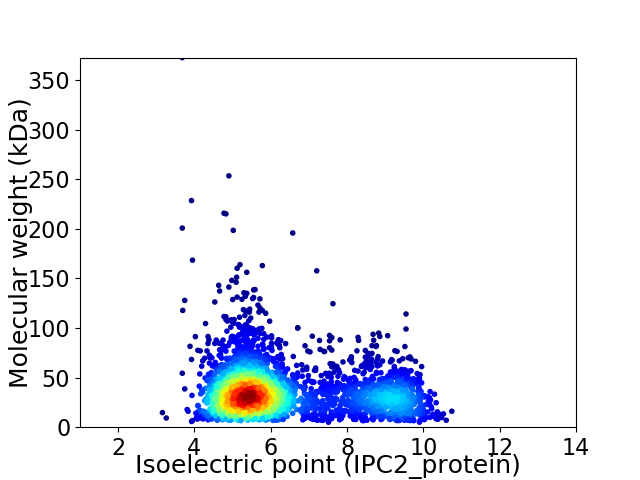
Virtual 2D-PAGE plot for 3080 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P6Y1Y8|A0A0P6Y1Y8_9CHLR Amidohydro-rel domain-containing protein OS=Levilinea saccharolytica OX=229921 GN=ADN01_17615 PE=4 SV=1
MM1 pKa = 7.12FAVSLLALIFFIGLAVDD18 pKa = 3.8AGAIYY23 pKa = 8.89VTYY26 pKa = 9.88GQLKK30 pKa = 9.07RR31 pKa = 11.84AVDD34 pKa = 3.75AASVAAANDD43 pKa = 3.58FKK45 pKa = 11.1KK46 pKa = 10.89GYY48 pKa = 8.43TRR50 pKa = 11.84DD51 pKa = 3.51QMEE54 pKa = 4.2DD55 pKa = 2.71SARR58 pKa = 11.84QVLIVHH64 pKa = 5.75QVDD67 pKa = 4.14LDD69 pKa = 4.38RR70 pKa = 11.84ITLNVYY76 pKa = 9.87ICDD79 pKa = 3.97NNGDD83 pKa = 3.92GARR86 pKa = 11.84DD87 pKa = 3.43ADD89 pKa = 4.08LQTNVPQFFARR100 pKa = 11.84CPDD103 pKa = 3.57TANGEE108 pKa = 4.38SPRR111 pKa = 11.84KK112 pKa = 9.16FVWVDD117 pKa = 3.06ASMKK121 pKa = 10.54APFYY125 pKa = 10.56FLSLLGFQSVDD136 pKa = 2.96LSTNAISEE144 pKa = 4.2AAPIDD149 pKa = 4.03LLIVLDD155 pKa = 4.04ISEE158 pKa = 4.43SMGVDD163 pKa = 3.27TPGYY167 pKa = 8.13TVNDD171 pKa = 4.24FNPANCNANTEE182 pKa = 4.51SYY184 pKa = 11.48DD185 pKa = 4.37DD186 pKa = 5.07DD187 pKa = 6.28DD188 pKa = 4.55NAATAAIPIVGNCAPLIQAKK208 pKa = 9.64KK209 pKa = 10.15AAIKK213 pKa = 10.69LVDD216 pKa = 3.86TLYY219 pKa = 11.06DD220 pKa = 3.94GYY222 pKa = 11.39DD223 pKa = 3.39SVSVVTFDD231 pKa = 3.17QVAYY235 pKa = 9.24QVPIADD241 pKa = 4.44TYY243 pKa = 12.24DD244 pKa = 3.19MAAVKK249 pKa = 9.93QAIGAIALHH258 pKa = 7.35DD259 pKa = 4.53DD260 pKa = 3.65PPYY263 pKa = 10.99NRR265 pKa = 11.84LWDD268 pKa = 3.35TWKK271 pKa = 10.96NPGRR275 pKa = 11.84VNLSNPEE282 pKa = 4.01DD283 pKa = 4.43RR284 pKa = 11.84DD285 pKa = 3.71GDD287 pKa = 3.86GADD290 pKa = 3.63YY291 pKa = 11.08DD292 pKa = 3.94NPAVVGYY299 pKa = 7.2TCPTFTANDD308 pKa = 3.89GTEE311 pKa = 4.01FGPPEE316 pKa = 4.05EE317 pKa = 5.45HH318 pKa = 7.18YY319 pKa = 11.31LDD321 pKa = 4.47DD322 pKa = 4.77RR323 pKa = 11.84WWQTGNPALVPGGEE337 pKa = 4.23GAPDD341 pKa = 3.65PFGWGGVPCDD351 pKa = 4.91RR352 pKa = 11.84DD353 pKa = 4.01DD354 pKa = 5.81KK355 pKa = 11.5LDD357 pKa = 3.9AYY359 pKa = 10.83DD360 pKa = 3.34WDD362 pKa = 5.16RR363 pKa = 11.84NGKK366 pKa = 5.66WTQYY370 pKa = 11.49DD371 pKa = 3.8HH372 pKa = 7.09DD373 pKa = 4.56TSGAWLTANDD383 pKa = 4.39PDD385 pKa = 4.16GAGALSVTLSPLSTCTGCGIRR406 pKa = 11.84VAANVLRR413 pKa = 11.84QTGRR417 pKa = 11.84PSSVWVMVLLSDD429 pKa = 4.49GAVNLSDD436 pKa = 3.2THH438 pKa = 7.63VNAGEE443 pKa = 4.07LNGEE447 pKa = 4.2DD448 pKa = 5.08VIPSWFPMGFCTGRR462 pKa = 11.84LFGPCVKK469 pKa = 10.55DD470 pKa = 3.83FGVGAEE476 pKa = 4.84DD477 pKa = 4.46GSANDD482 pKa = 5.49GYY484 pKa = 10.31WCSACQDD491 pKa = 3.3NFKK494 pKa = 9.97ATRR497 pKa = 11.84YY498 pKa = 9.93CVDD501 pKa = 3.97HH502 pKa = 7.22EE503 pKa = 4.47EE504 pKa = 4.27ATCPDD509 pKa = 3.69DD510 pKa = 3.78TTWIGDD516 pKa = 3.18TWTPNTLQKK525 pKa = 10.42RR526 pKa = 11.84YY527 pKa = 9.97SVYY530 pKa = 10.76DD531 pKa = 3.45YY532 pKa = 11.33ALDD535 pKa = 3.79MTDD538 pKa = 3.12EE539 pKa = 4.48AALTRR544 pKa = 11.84STKK547 pKa = 9.42LTEE550 pKa = 3.91PAGNDD555 pKa = 2.77IAIYY559 pKa = 9.65TIGLGAASSGADD571 pKa = 3.44LLRR574 pKa = 11.84YY575 pKa = 9.0LAAVGDD581 pKa = 4.36DD582 pKa = 3.92GDD584 pKa = 4.15RR585 pKa = 11.84TTDD588 pKa = 3.04PCAPFAANPTRR599 pKa = 11.84DD600 pKa = 3.1CGQYY604 pKa = 10.13YY605 pKa = 9.73FADD608 pKa = 3.44TSYY611 pKa = 10.71EE612 pKa = 3.89LQEE615 pKa = 4.07IFEE618 pKa = 5.66DD619 pKa = 3.39IASRR623 pKa = 11.84IYY625 pKa = 10.65TRR627 pKa = 11.84ITDD630 pKa = 3.36
MM1 pKa = 7.12FAVSLLALIFFIGLAVDD18 pKa = 3.8AGAIYY23 pKa = 8.89VTYY26 pKa = 9.88GQLKK30 pKa = 9.07RR31 pKa = 11.84AVDD34 pKa = 3.75AASVAAANDD43 pKa = 3.58FKK45 pKa = 11.1KK46 pKa = 10.89GYY48 pKa = 8.43TRR50 pKa = 11.84DD51 pKa = 3.51QMEE54 pKa = 4.2DD55 pKa = 2.71SARR58 pKa = 11.84QVLIVHH64 pKa = 5.75QVDD67 pKa = 4.14LDD69 pKa = 4.38RR70 pKa = 11.84ITLNVYY76 pKa = 9.87ICDD79 pKa = 3.97NNGDD83 pKa = 3.92GARR86 pKa = 11.84DD87 pKa = 3.43ADD89 pKa = 4.08LQTNVPQFFARR100 pKa = 11.84CPDD103 pKa = 3.57TANGEE108 pKa = 4.38SPRR111 pKa = 11.84KK112 pKa = 9.16FVWVDD117 pKa = 3.06ASMKK121 pKa = 10.54APFYY125 pKa = 10.56FLSLLGFQSVDD136 pKa = 2.96LSTNAISEE144 pKa = 4.2AAPIDD149 pKa = 4.03LLIVLDD155 pKa = 4.04ISEE158 pKa = 4.43SMGVDD163 pKa = 3.27TPGYY167 pKa = 8.13TVNDD171 pKa = 4.24FNPANCNANTEE182 pKa = 4.51SYY184 pKa = 11.48DD185 pKa = 4.37DD186 pKa = 5.07DD187 pKa = 6.28DD188 pKa = 4.55NAATAAIPIVGNCAPLIQAKK208 pKa = 9.64KK209 pKa = 10.15AAIKK213 pKa = 10.69LVDD216 pKa = 3.86TLYY219 pKa = 11.06DD220 pKa = 3.94GYY222 pKa = 11.39DD223 pKa = 3.39SVSVVTFDD231 pKa = 3.17QVAYY235 pKa = 9.24QVPIADD241 pKa = 4.44TYY243 pKa = 12.24DD244 pKa = 3.19MAAVKK249 pKa = 9.93QAIGAIALHH258 pKa = 7.35DD259 pKa = 4.53DD260 pKa = 3.65PPYY263 pKa = 10.99NRR265 pKa = 11.84LWDD268 pKa = 3.35TWKK271 pKa = 10.96NPGRR275 pKa = 11.84VNLSNPEE282 pKa = 4.01DD283 pKa = 4.43RR284 pKa = 11.84DD285 pKa = 3.71GDD287 pKa = 3.86GADD290 pKa = 3.63YY291 pKa = 11.08DD292 pKa = 3.94NPAVVGYY299 pKa = 7.2TCPTFTANDD308 pKa = 3.89GTEE311 pKa = 4.01FGPPEE316 pKa = 4.05EE317 pKa = 5.45HH318 pKa = 7.18YY319 pKa = 11.31LDD321 pKa = 4.47DD322 pKa = 4.77RR323 pKa = 11.84WWQTGNPALVPGGEE337 pKa = 4.23GAPDD341 pKa = 3.65PFGWGGVPCDD351 pKa = 4.91RR352 pKa = 11.84DD353 pKa = 4.01DD354 pKa = 5.81KK355 pKa = 11.5LDD357 pKa = 3.9AYY359 pKa = 10.83DD360 pKa = 3.34WDD362 pKa = 5.16RR363 pKa = 11.84NGKK366 pKa = 5.66WTQYY370 pKa = 11.49DD371 pKa = 3.8HH372 pKa = 7.09DD373 pKa = 4.56TSGAWLTANDD383 pKa = 4.39PDD385 pKa = 4.16GAGALSVTLSPLSTCTGCGIRR406 pKa = 11.84VAANVLRR413 pKa = 11.84QTGRR417 pKa = 11.84PSSVWVMVLLSDD429 pKa = 4.49GAVNLSDD436 pKa = 3.2THH438 pKa = 7.63VNAGEE443 pKa = 4.07LNGEE447 pKa = 4.2DD448 pKa = 5.08VIPSWFPMGFCTGRR462 pKa = 11.84LFGPCVKK469 pKa = 10.55DD470 pKa = 3.83FGVGAEE476 pKa = 4.84DD477 pKa = 4.46GSANDD482 pKa = 5.49GYY484 pKa = 10.31WCSACQDD491 pKa = 3.3NFKK494 pKa = 9.97ATRR497 pKa = 11.84YY498 pKa = 9.93CVDD501 pKa = 3.97HH502 pKa = 7.22EE503 pKa = 4.47EE504 pKa = 4.27ATCPDD509 pKa = 3.69DD510 pKa = 3.78TTWIGDD516 pKa = 3.18TWTPNTLQKK525 pKa = 10.42RR526 pKa = 11.84YY527 pKa = 9.97SVYY530 pKa = 10.76DD531 pKa = 3.45YY532 pKa = 11.33ALDD535 pKa = 3.79MTDD538 pKa = 3.12EE539 pKa = 4.48AALTRR544 pKa = 11.84STKK547 pKa = 9.42LTEE550 pKa = 3.91PAGNDD555 pKa = 2.77IAIYY559 pKa = 9.65TIGLGAASSGADD571 pKa = 3.44LLRR574 pKa = 11.84YY575 pKa = 9.0LAAVGDD581 pKa = 4.36DD582 pKa = 3.92GDD584 pKa = 4.15RR585 pKa = 11.84TTDD588 pKa = 3.04PCAPFAANPTRR599 pKa = 11.84DD600 pKa = 3.1CGQYY604 pKa = 10.13YY605 pKa = 9.73FADD608 pKa = 3.44TSYY611 pKa = 10.71EE612 pKa = 3.89LQEE615 pKa = 4.07IFEE618 pKa = 5.66DD619 pKa = 3.39IASRR623 pKa = 11.84IYY625 pKa = 10.65TRR627 pKa = 11.84ITDD630 pKa = 3.36
Molecular weight: 68.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M9U2T0|A0A0M9U2T0_9CHLR ABC transporter permease OS=Levilinea saccharolytica OX=229921 GN=ADN01_16740 PE=4 SV=1
MM1 pKa = 7.97DD2 pKa = 5.05PRR4 pKa = 11.84WYY6 pKa = 10.1PLILFVALLLLWGVLNAAFRR26 pKa = 11.84RR27 pKa = 11.84HH28 pKa = 5.32RR29 pKa = 11.84QEE31 pKa = 3.87DD32 pKa = 4.18CPRR35 pKa = 11.84CGGPIEE41 pKa = 4.75RR42 pKa = 11.84IHH44 pKa = 7.06RR45 pKa = 11.84SKK47 pKa = 11.15LEE49 pKa = 3.84KK50 pKa = 10.69ALGLLLVYY58 pKa = 7.84PTGKK62 pKa = 9.63YY63 pKa = 9.21QCDD66 pKa = 3.08QCGWIGLRR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84SHH78 pKa = 6.07RR79 pKa = 11.84RR80 pKa = 11.84STRR83 pKa = 11.84TT84 pKa = 3.07
MM1 pKa = 7.97DD2 pKa = 5.05PRR4 pKa = 11.84WYY6 pKa = 10.1PLILFVALLLLWGVLNAAFRR26 pKa = 11.84RR27 pKa = 11.84HH28 pKa = 5.32RR29 pKa = 11.84QEE31 pKa = 3.87DD32 pKa = 4.18CPRR35 pKa = 11.84CGGPIEE41 pKa = 4.75RR42 pKa = 11.84IHH44 pKa = 7.06RR45 pKa = 11.84SKK47 pKa = 11.15LEE49 pKa = 3.84KK50 pKa = 10.69ALGLLLVYY58 pKa = 7.84PTGKK62 pKa = 9.63YY63 pKa = 9.21QCDD66 pKa = 3.08QCGWIGLRR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84SHH78 pKa = 6.07RR79 pKa = 11.84RR80 pKa = 11.84STRR83 pKa = 11.84TT84 pKa = 3.07
Molecular weight: 9.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1055798 |
46 |
3580 |
342.8 |
37.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.801 ± 0.061 | 0.825 ± 0.012 |
4.801 ± 0.033 | 5.925 ± 0.052 |
3.908 ± 0.03 | 7.923 ± 0.044 |
1.885 ± 0.021 | 5.029 ± 0.037 |
2.997 ± 0.036 | 11.486 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.399 ± 0.023 | 2.948 ± 0.024 |
5.546 ± 0.037 | 4.31 ± 0.031 |
6.354 ± 0.044 | 5.443 ± 0.031 |
5.335 ± 0.042 | 7.558 ± 0.044 |
1.691 ± 0.024 | 2.836 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
