
Microdochium bolleyi
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Xylariomycetidae; Xylariales; Microdochiaceae; Microdochium
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
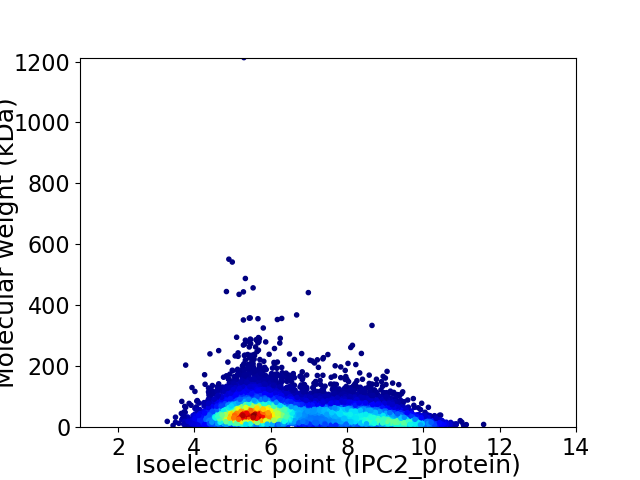
Virtual 2D-PAGE plot for 13164 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A136IME4|A0A136IME4_9PEZI S-adenosylmethionine synthase OS=Microdochium bolleyi OX=196109 GN=Micbo1qcDRAFT_140634 PE=3 SV=1
MM1 pKa = 7.94PLNNHH6 pKa = 5.49NQRR9 pKa = 11.84LEE11 pKa = 4.19KK12 pKa = 10.1TMHH15 pKa = 6.19TISYY19 pKa = 9.94ASNNSYY25 pKa = 10.29FRR27 pKa = 11.84DD28 pKa = 3.71VQHH31 pKa = 7.35GSRR34 pKa = 11.84TEE36 pKa = 4.14SNTKK40 pKa = 9.77IAIEE44 pKa = 4.39TSTQAQPGSPADD56 pKa = 4.36HH57 pKa = 6.74ILVPLNTGDD66 pKa = 3.65QLLSNDD72 pKa = 3.44VATEE76 pKa = 3.99DD77 pKa = 3.88TMSPGAPEE85 pKa = 4.08HH86 pKa = 7.12CEE88 pKa = 3.84DD89 pKa = 4.04QAEE92 pKa = 4.26TEE94 pKa = 4.26PLSVHH99 pKa = 5.78TSIGLQTPGNAEE111 pKa = 3.71TSMLNTAQNPGQDD124 pKa = 3.82PPQDD128 pKa = 4.03PLQSPPQHH136 pKa = 6.58LAQNPGHH143 pKa = 6.42ASGYY147 pKa = 10.33ISDD150 pKa = 3.97TAQANARR157 pKa = 11.84PPEE160 pKa = 4.32PFQQATAQGEE170 pKa = 4.39INSNHH175 pKa = 6.95DD176 pKa = 3.15EE177 pKa = 4.61SALQTYY183 pKa = 8.01NQSQHH188 pKa = 6.16HH189 pKa = 5.63SQNTNHH195 pKa = 6.71PQQFSVRR202 pKa = 11.84QEE204 pKa = 4.02EE205 pKa = 4.68PGTSSAPAKK214 pKa = 10.25KK215 pKa = 10.14SSRR218 pKa = 11.84TDD220 pKa = 2.98NLLALGAGMVAGAAIGAAGYY240 pKa = 9.51AAYY243 pKa = 9.68KK244 pKa = 9.98KK245 pKa = 10.55LQGDD249 pKa = 3.92GEE251 pKa = 4.53GRR253 pKa = 11.84VQDD256 pKa = 5.47ARR258 pKa = 11.84DD259 pKa = 3.59SDD261 pKa = 4.07NDD263 pKa = 3.92SVLSWVSSISSRR275 pKa = 11.84SHH277 pKa = 6.81SSEE280 pKa = 3.97PNSVAGQGDD289 pKa = 4.11QMEE292 pKa = 4.84LDD294 pKa = 4.05YY295 pKa = 11.48EE296 pKa = 4.29QGTDD300 pKa = 3.26AQTKK304 pKa = 10.3SEE306 pKa = 4.58GFYY309 pKa = 10.87EE310 pKa = 4.39DD311 pKa = 5.64QFTSWDD317 pKa = 3.48QYY319 pKa = 11.63NEE321 pKa = 3.95YY322 pKa = 10.52QVFPEE327 pKa = 4.33EE328 pKa = 4.21VGSRR332 pKa = 11.84SYY334 pKa = 11.27GGPDD338 pKa = 3.44DD339 pKa = 5.19EE340 pKa = 5.64SWTSDD345 pKa = 3.5LPSDD349 pKa = 3.86AGSKK353 pKa = 10.17RR354 pKa = 11.84GISPAASLYY363 pKa = 10.89DD364 pKa = 3.6EE365 pKa = 4.36NEE367 pKa = 3.77YY368 pKa = 9.71WRR370 pKa = 11.84NSDD373 pKa = 3.78PQDD376 pKa = 3.86PDD378 pKa = 3.25VKK380 pKa = 10.72EE381 pKa = 4.28VMSGSAQHH389 pKa = 6.43SPISLSDD396 pKa = 3.52YY397 pKa = 10.1SAHH400 pKa = 6.57GLQSPGIVQPFDD412 pKa = 3.05WEE414 pKa = 4.55TNSDD418 pKa = 3.59QNGDD422 pKa = 3.26HH423 pKa = 7.12GDD425 pKa = 3.51PHH427 pKa = 8.22RR428 pKa = 11.84SVPPGSGEE436 pKa = 3.95DD437 pKa = 3.68HH438 pKa = 7.42DD439 pKa = 4.87VSDD442 pKa = 4.22WGQGDD447 pKa = 4.01GSVDD451 pKa = 3.69GSVHH455 pKa = 6.48GYY457 pKa = 9.91DD458 pKa = 5.03RR459 pKa = 11.84EE460 pKa = 4.22YY461 pKa = 10.69PDD463 pKa = 4.16YY464 pKa = 11.17DD465 pKa = 3.47PEE467 pKa = 4.24YY468 pKa = 10.38LHH470 pKa = 6.87SQFGDD475 pKa = 3.62EE476 pKa = 4.99LGSDD480 pKa = 3.75ADD482 pKa = 3.94SAAGALSEE490 pKa = 4.86VEE492 pKa = 3.89PSEE495 pKa = 4.3NGSALSGDD503 pKa = 4.24EE504 pKa = 4.83DD505 pKa = 5.01DD506 pKa = 6.6LMSQRR511 pKa = 11.84DD512 pKa = 3.56FSHH515 pKa = 7.32AYY517 pKa = 9.97EE518 pKa = 5.12SDD520 pKa = 3.88DD521 pKa = 4.68NPGDD525 pKa = 3.57EE526 pKa = 5.24DD527 pKa = 4.09GSYY530 pKa = 11.08SPGYY534 pKa = 10.68DD535 pKa = 3.5SDD537 pKa = 3.87AQSDD541 pKa = 3.93GFAGYY546 pKa = 10.56GSQRR550 pKa = 11.84DD551 pKa = 3.8EE552 pKa = 4.42YY553 pKa = 11.35GAGDD557 pKa = 5.12NIDD560 pKa = 5.16DD561 pKa = 5.36DD562 pKa = 5.2DD563 pKa = 6.16DD564 pKa = 5.62SGEE567 pKa = 4.21GEE569 pKa = 3.88QAYY572 pKa = 10.14EE573 pKa = 4.43SGGDD577 pKa = 3.89DD578 pKa = 4.11GGDD581 pKa = 3.79GNDD584 pKa = 3.44EE585 pKa = 4.46SQGEE589 pKa = 4.28AYY591 pKa = 10.13GYY593 pKa = 10.85DD594 pKa = 3.63GAQDD598 pKa = 3.69NPEE601 pKa = 3.98GAEE604 pKa = 4.05DD605 pKa = 3.7SGEE608 pKa = 4.18DD609 pKa = 3.79EE610 pKa = 5.31QIDD613 pKa = 4.22GPDD616 pKa = 3.51YY617 pKa = 11.11EE618 pKa = 5.09GNDD621 pKa = 3.95YY622 pKa = 11.63GDD624 pKa = 4.33ADD626 pKa = 4.68YY627 pKa = 11.75GDD629 pKa = 5.47DD630 pKa = 4.67DD631 pKa = 5.18DD632 pKa = 6.27GFYY635 pKa = 11.36EE636 pKa = 4.3EE637 pKa = 4.99
MM1 pKa = 7.94PLNNHH6 pKa = 5.49NQRR9 pKa = 11.84LEE11 pKa = 4.19KK12 pKa = 10.1TMHH15 pKa = 6.19TISYY19 pKa = 9.94ASNNSYY25 pKa = 10.29FRR27 pKa = 11.84DD28 pKa = 3.71VQHH31 pKa = 7.35GSRR34 pKa = 11.84TEE36 pKa = 4.14SNTKK40 pKa = 9.77IAIEE44 pKa = 4.39TSTQAQPGSPADD56 pKa = 4.36HH57 pKa = 6.74ILVPLNTGDD66 pKa = 3.65QLLSNDD72 pKa = 3.44VATEE76 pKa = 3.99DD77 pKa = 3.88TMSPGAPEE85 pKa = 4.08HH86 pKa = 7.12CEE88 pKa = 3.84DD89 pKa = 4.04QAEE92 pKa = 4.26TEE94 pKa = 4.26PLSVHH99 pKa = 5.78TSIGLQTPGNAEE111 pKa = 3.71TSMLNTAQNPGQDD124 pKa = 3.82PPQDD128 pKa = 4.03PLQSPPQHH136 pKa = 6.58LAQNPGHH143 pKa = 6.42ASGYY147 pKa = 10.33ISDD150 pKa = 3.97TAQANARR157 pKa = 11.84PPEE160 pKa = 4.32PFQQATAQGEE170 pKa = 4.39INSNHH175 pKa = 6.95DD176 pKa = 3.15EE177 pKa = 4.61SALQTYY183 pKa = 8.01NQSQHH188 pKa = 6.16HH189 pKa = 5.63SQNTNHH195 pKa = 6.71PQQFSVRR202 pKa = 11.84QEE204 pKa = 4.02EE205 pKa = 4.68PGTSSAPAKK214 pKa = 10.25KK215 pKa = 10.14SSRR218 pKa = 11.84TDD220 pKa = 2.98NLLALGAGMVAGAAIGAAGYY240 pKa = 9.51AAYY243 pKa = 9.68KK244 pKa = 9.98KK245 pKa = 10.55LQGDD249 pKa = 3.92GEE251 pKa = 4.53GRR253 pKa = 11.84VQDD256 pKa = 5.47ARR258 pKa = 11.84DD259 pKa = 3.59SDD261 pKa = 4.07NDD263 pKa = 3.92SVLSWVSSISSRR275 pKa = 11.84SHH277 pKa = 6.81SSEE280 pKa = 3.97PNSVAGQGDD289 pKa = 4.11QMEE292 pKa = 4.84LDD294 pKa = 4.05YY295 pKa = 11.48EE296 pKa = 4.29QGTDD300 pKa = 3.26AQTKK304 pKa = 10.3SEE306 pKa = 4.58GFYY309 pKa = 10.87EE310 pKa = 4.39DD311 pKa = 5.64QFTSWDD317 pKa = 3.48QYY319 pKa = 11.63NEE321 pKa = 3.95YY322 pKa = 10.52QVFPEE327 pKa = 4.33EE328 pKa = 4.21VGSRR332 pKa = 11.84SYY334 pKa = 11.27GGPDD338 pKa = 3.44DD339 pKa = 5.19EE340 pKa = 5.64SWTSDD345 pKa = 3.5LPSDD349 pKa = 3.86AGSKK353 pKa = 10.17RR354 pKa = 11.84GISPAASLYY363 pKa = 10.89DD364 pKa = 3.6EE365 pKa = 4.36NEE367 pKa = 3.77YY368 pKa = 9.71WRR370 pKa = 11.84NSDD373 pKa = 3.78PQDD376 pKa = 3.86PDD378 pKa = 3.25VKK380 pKa = 10.72EE381 pKa = 4.28VMSGSAQHH389 pKa = 6.43SPISLSDD396 pKa = 3.52YY397 pKa = 10.1SAHH400 pKa = 6.57GLQSPGIVQPFDD412 pKa = 3.05WEE414 pKa = 4.55TNSDD418 pKa = 3.59QNGDD422 pKa = 3.26HH423 pKa = 7.12GDD425 pKa = 3.51PHH427 pKa = 8.22RR428 pKa = 11.84SVPPGSGEE436 pKa = 3.95DD437 pKa = 3.68HH438 pKa = 7.42DD439 pKa = 4.87VSDD442 pKa = 4.22WGQGDD447 pKa = 4.01GSVDD451 pKa = 3.69GSVHH455 pKa = 6.48GYY457 pKa = 9.91DD458 pKa = 5.03RR459 pKa = 11.84EE460 pKa = 4.22YY461 pKa = 10.69PDD463 pKa = 4.16YY464 pKa = 11.17DD465 pKa = 3.47PEE467 pKa = 4.24YY468 pKa = 10.38LHH470 pKa = 6.87SQFGDD475 pKa = 3.62EE476 pKa = 4.99LGSDD480 pKa = 3.75ADD482 pKa = 3.94SAAGALSEE490 pKa = 4.86VEE492 pKa = 3.89PSEE495 pKa = 4.3NGSALSGDD503 pKa = 4.24EE504 pKa = 4.83DD505 pKa = 5.01DD506 pKa = 6.6LMSQRR511 pKa = 11.84DD512 pKa = 3.56FSHH515 pKa = 7.32AYY517 pKa = 9.97EE518 pKa = 5.12SDD520 pKa = 3.88DD521 pKa = 4.68NPGDD525 pKa = 3.57EE526 pKa = 5.24DD527 pKa = 4.09GSYY530 pKa = 11.08SPGYY534 pKa = 10.68DD535 pKa = 3.5SDD537 pKa = 3.87AQSDD541 pKa = 3.93GFAGYY546 pKa = 10.56GSQRR550 pKa = 11.84DD551 pKa = 3.8EE552 pKa = 4.42YY553 pKa = 11.35GAGDD557 pKa = 5.12NIDD560 pKa = 5.16DD561 pKa = 5.36DD562 pKa = 5.2DD563 pKa = 6.16DD564 pKa = 5.62SGEE567 pKa = 4.21GEE569 pKa = 3.88QAYY572 pKa = 10.14EE573 pKa = 4.43SGGDD577 pKa = 3.89DD578 pKa = 4.11GGDD581 pKa = 3.79GNDD584 pKa = 3.44EE585 pKa = 4.46SQGEE589 pKa = 4.28AYY591 pKa = 10.13GYY593 pKa = 10.85DD594 pKa = 3.63GAQDD598 pKa = 3.69NPEE601 pKa = 3.98GAEE604 pKa = 4.05DD605 pKa = 3.7SGEE608 pKa = 4.18DD609 pKa = 3.79EE610 pKa = 5.31QIDD613 pKa = 4.22GPDD616 pKa = 3.51YY617 pKa = 11.11EE618 pKa = 5.09GNDD621 pKa = 3.95YY622 pKa = 11.63GDD624 pKa = 4.33ADD626 pKa = 4.68YY627 pKa = 11.75GDD629 pKa = 5.47DD630 pKa = 4.67DD631 pKa = 5.18DD632 pKa = 6.27GFYY635 pKa = 11.36EE636 pKa = 4.3EE637 pKa = 4.99
Molecular weight: 68.27 kDa
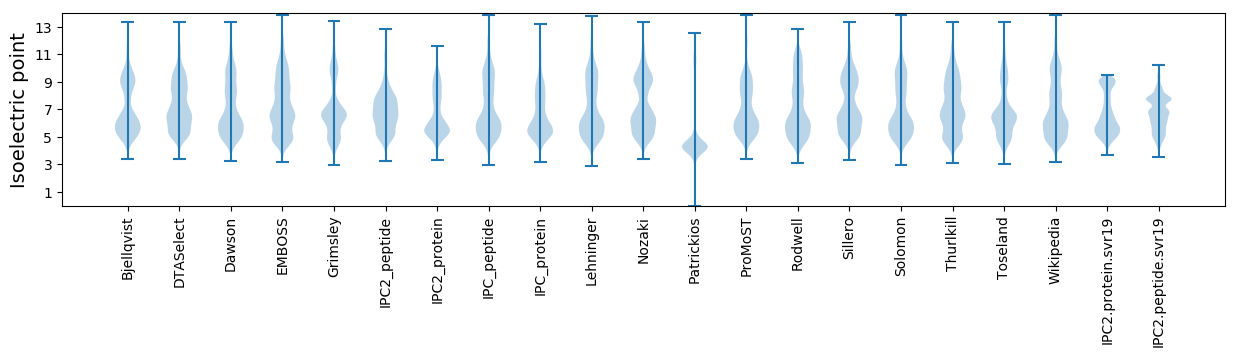
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A136IRZ4|A0A136IRZ4_9PEZI Uncharacterized protein (Fragment) OS=Microdochium bolleyi OX=196109 GN=Micbo1qcDRAFT_167258 PE=4 SV=1
MM1 pKa = 7.71ARR3 pKa = 11.84TTLLHH8 pKa = 6.24RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84IRR13 pKa = 11.84LLTLGAGPVAWRR25 pKa = 11.84WPASSVEE32 pKa = 3.86ARR34 pKa = 11.84RR35 pKa = 11.84GTRR38 pKa = 11.84RR39 pKa = 11.84QAGTRR44 pKa = 11.84RR45 pKa = 11.84GVRR48 pKa = 11.84AGRR51 pKa = 11.84PAGRR55 pKa = 11.84HH56 pKa = 3.79RR57 pKa = 11.84HH58 pKa = 4.87RR59 pKa = 11.84SARR62 pKa = 11.84FAQGPGSPAGGKK74 pKa = 7.41QVV76 pKa = 3.01
MM1 pKa = 7.71ARR3 pKa = 11.84TTLLHH8 pKa = 6.24RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84IRR13 pKa = 11.84LLTLGAGPVAWRR25 pKa = 11.84WPASSVEE32 pKa = 3.86ARR34 pKa = 11.84RR35 pKa = 11.84GTRR38 pKa = 11.84RR39 pKa = 11.84QAGTRR44 pKa = 11.84RR45 pKa = 11.84GVRR48 pKa = 11.84AGRR51 pKa = 11.84PAGRR55 pKa = 11.84HH56 pKa = 3.79RR57 pKa = 11.84HH58 pKa = 4.87RR59 pKa = 11.84SARR62 pKa = 11.84FAQGPGSPAGGKK74 pKa = 7.41QVV76 pKa = 3.01
Molecular weight: 8.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5816858 |
50 |
11096 |
441.9 |
48.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.748 ± 0.022 | 1.32 ± 0.009 |
5.777 ± 0.017 | 5.687 ± 0.019 |
3.557 ± 0.013 | 7.374 ± 0.02 |
2.482 ± 0.011 | 4.485 ± 0.017 |
4.373 ± 0.021 | 8.777 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.226 ± 0.007 | 3.318 ± 0.012 |
6.013 ± 0.019 | 4.008 ± 0.016 |
6.29 ± 0.018 | 8.048 ± 0.023 |
6.079 ± 0.021 | 6.304 ± 0.017 |
1.509 ± 0.008 | 2.627 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
