
Nerine virus X
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Alphaflexiviridae; Potexvirus
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
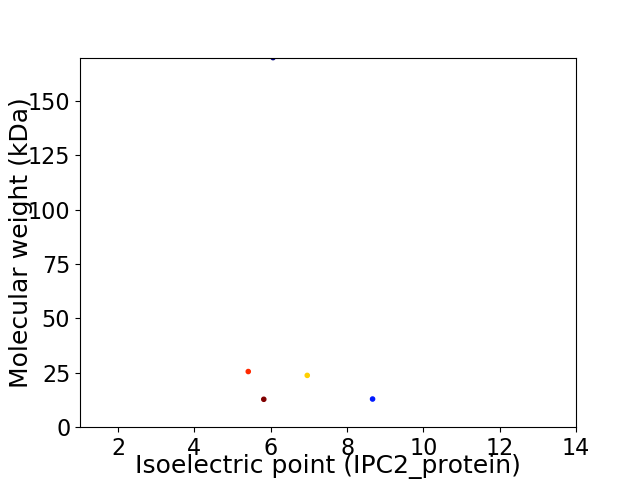
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q2V0S0|Q2V0S0_9VIRU ORF1 protein OS=Nerine virus X OX=333348 PE=4 SV=1
MM1 pKa = 7.5EE2 pKa = 5.7NIINKK7 pKa = 8.91LQTRR11 pKa = 11.84FTRR14 pKa = 11.84TSLPISKK21 pKa = 9.66PIIIHH26 pKa = 5.93CCAGAGKK33 pKa = 7.33TTFIRR38 pKa = 11.84EE39 pKa = 4.2LLEE42 pKa = 3.95QFPEE46 pKa = 4.35VQAYY50 pKa = 6.81THH52 pKa = 5.73SQHH55 pKa = 6.61LAAEE59 pKa = 4.35KK60 pKa = 9.88TISGRR65 pKa = 11.84NFHH68 pKa = 6.82HH69 pKa = 6.9FSEE72 pKa = 4.55YY73 pKa = 9.67QPGYY77 pKa = 10.42LDD79 pKa = 5.39VIDD82 pKa = 5.35EE83 pKa = 4.35YY84 pKa = 11.62LAGPIPEE91 pKa = 4.28TCHH94 pKa = 6.21FCFADD99 pKa = 4.76PYY101 pKa = 10.75QYY103 pKa = 11.3NIDD106 pKa = 3.98ALPAHH111 pKa = 6.96FICNKK116 pKa = 9.37SYY118 pKa = 11.23RR119 pKa = 11.84FGTLTADD126 pKa = 3.67YY127 pKa = 9.97LNSLGYY133 pKa = 8.14QVRR136 pKa = 11.84SAGGSDD142 pKa = 3.77SIRR145 pKa = 11.84FIDD148 pKa = 3.88LAHH151 pKa = 7.06WEE153 pKa = 4.3PEE155 pKa = 4.83GICITQDD162 pKa = 3.06SDD164 pKa = 3.85ILNLLTRR171 pKa = 11.84HH172 pKa = 6.65GISAYY177 pKa = 10.19HH178 pKa = 6.1PCEE181 pKa = 3.81TLGLQFNCVDD191 pKa = 3.61YY192 pKa = 10.86LVPKK196 pKa = 10.55LPTPATADD204 pKa = 3.34ANTYY208 pKa = 10.17LALTRR213 pKa = 11.84HH214 pKa = 5.53SSRR217 pKa = 11.84LNVVSDD223 pKa = 3.93AADD226 pKa = 3.31SATT229 pKa = 3.56
MM1 pKa = 7.5EE2 pKa = 5.7NIINKK7 pKa = 8.91LQTRR11 pKa = 11.84FTRR14 pKa = 11.84TSLPISKK21 pKa = 9.66PIIIHH26 pKa = 5.93CCAGAGKK33 pKa = 7.33TTFIRR38 pKa = 11.84EE39 pKa = 4.2LLEE42 pKa = 3.95QFPEE46 pKa = 4.35VQAYY50 pKa = 6.81THH52 pKa = 5.73SQHH55 pKa = 6.61LAAEE59 pKa = 4.35KK60 pKa = 9.88TISGRR65 pKa = 11.84NFHH68 pKa = 6.82HH69 pKa = 6.9FSEE72 pKa = 4.55YY73 pKa = 9.67QPGYY77 pKa = 10.42LDD79 pKa = 5.39VIDD82 pKa = 5.35EE83 pKa = 4.35YY84 pKa = 11.62LAGPIPEE91 pKa = 4.28TCHH94 pKa = 6.21FCFADD99 pKa = 4.76PYY101 pKa = 10.75QYY103 pKa = 11.3NIDD106 pKa = 3.98ALPAHH111 pKa = 6.96FICNKK116 pKa = 9.37SYY118 pKa = 11.23RR119 pKa = 11.84FGTLTADD126 pKa = 3.67YY127 pKa = 9.97LNSLGYY133 pKa = 8.14QVRR136 pKa = 11.84SAGGSDD142 pKa = 3.77SIRR145 pKa = 11.84FIDD148 pKa = 3.88LAHH151 pKa = 7.06WEE153 pKa = 4.3PEE155 pKa = 4.83GICITQDD162 pKa = 3.06SDD164 pKa = 3.85ILNLLTRR171 pKa = 11.84HH172 pKa = 6.65GISAYY177 pKa = 10.19HH178 pKa = 6.1PCEE181 pKa = 3.81TLGLQFNCVDD191 pKa = 3.61YY192 pKa = 10.86LVPKK196 pKa = 10.55LPTPATADD204 pKa = 3.34ANTYY208 pKa = 10.17LALTRR213 pKa = 11.84HH214 pKa = 5.53SSRR217 pKa = 11.84LNVVSDD223 pKa = 3.93AADD226 pKa = 3.31SATT229 pKa = 3.56
Molecular weight: 25.59 kDa
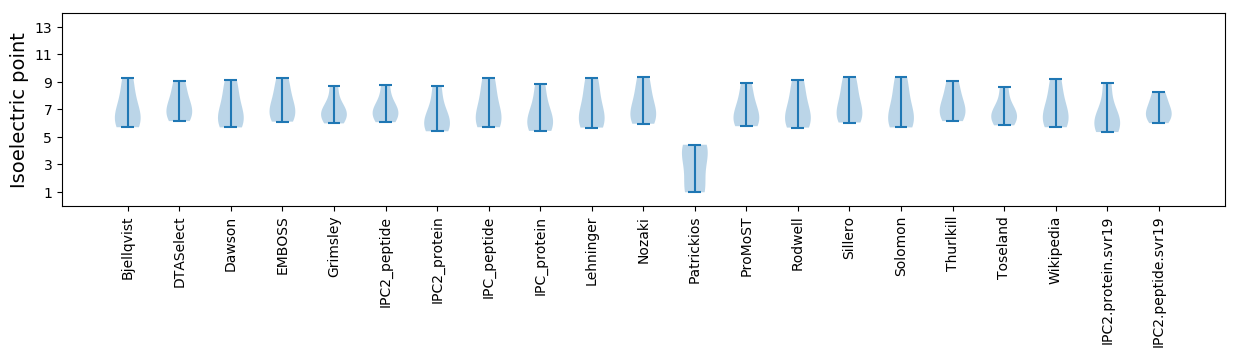
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q2V0R9|Q2V0R9_9VIRU Triple geneblock protein 1 OS=Nerine virus X OX=333348 PE=4 SV=1
MM1 pKa = 7.91PLTAPPDD8 pKa = 3.67YY9 pKa = 10.53THH11 pKa = 7.51ILPIAIVSIAVALSLYY27 pKa = 9.97TITRR31 pKa = 11.84NTLPHH36 pKa = 6.23TGDD39 pKa = 4.17NIHH42 pKa = 6.69HH43 pKa = 6.66FPHH46 pKa = 6.73GGRR49 pKa = 11.84YY50 pKa = 8.78RR51 pKa = 11.84DD52 pKa = 3.66GTKK55 pKa = 10.39SITYY59 pKa = 9.13CPPQRR64 pKa = 11.84NLSPFNSTGYY74 pKa = 10.48SIIPTALAILLPAAIYY90 pKa = 10.46LSSKK94 pKa = 9.81CFNSRR99 pKa = 11.84THH101 pKa = 6.05PHH103 pKa = 6.03SCSHH107 pKa = 6.31CQPNSATMRR116 pKa = 11.84GTSS119 pKa = 3.03
MM1 pKa = 7.91PLTAPPDD8 pKa = 3.67YY9 pKa = 10.53THH11 pKa = 7.51ILPIAIVSIAVALSLYY27 pKa = 9.97TITRR31 pKa = 11.84NTLPHH36 pKa = 6.23TGDD39 pKa = 4.17NIHH42 pKa = 6.69HH43 pKa = 6.66FPHH46 pKa = 6.73GGRR49 pKa = 11.84YY50 pKa = 8.78RR51 pKa = 11.84DD52 pKa = 3.66GTKK55 pKa = 10.39SITYY59 pKa = 9.13CPPQRR64 pKa = 11.84NLSPFNSTGYY74 pKa = 10.48SIIPTALAILLPAAIYY90 pKa = 10.46LSSKK94 pKa = 9.81CFNSRR99 pKa = 11.84THH101 pKa = 6.05PHH103 pKa = 6.03SCSHH107 pKa = 6.31CQPNSATMRR116 pKa = 11.84GTSS119 pKa = 3.03
Molecular weight: 12.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2193 |
119 |
1504 |
438.6 |
49.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.664 ± 1.74 | 1.87 ± 0.499 |
4.514 ± 0.583 | 5.928 ± 1.336 |
3.694 ± 0.47 | 4.423 ± 0.225 |
4.15 ± 0.564 | 6.065 ± 0.76 |
5.016 ± 1.123 | 9.166 ± 0.226 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.006 ± 0.387 | 4.606 ± 0.121 |
7.342 ± 0.729 | 4.514 ± 0.487 |
4.514 ± 0.271 | 7.068 ± 1.039 |
7.478 ± 0.568 | 4.651 ± 0.676 |
1.003 ± 0.258 | 3.329 ± 0.519 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
