
Cucumber mosaic virus (strain FNY) (CMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Cucumovirus; Cucumber mosaic virus
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
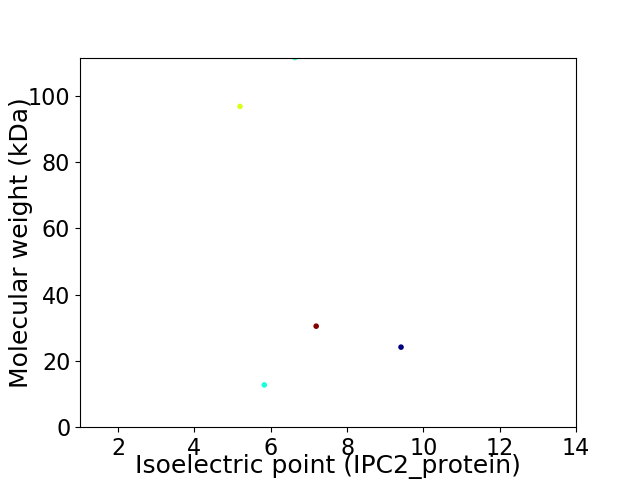
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P17769|1A_CMVFN Replication protein 1a OS=Cucumber mosaic virus (strain FNY) OX=12307 GN=ORF1a PE=3 SV=2
MM1 pKa = 7.78AFPAPAFSLANLLNGSYY18 pKa = 10.84GVDD21 pKa = 3.06TPEE24 pKa = 3.93DD25 pKa = 3.67VEE27 pKa = 4.3RR28 pKa = 11.84LRR30 pKa = 11.84SEE32 pKa = 4.02QRR34 pKa = 11.84EE35 pKa = 4.09EE36 pKa = 3.47AAAACRR42 pKa = 11.84NYY44 pKa = 10.59RR45 pKa = 11.84PLPAVDD51 pKa = 3.51VSEE54 pKa = 4.73SVTEE58 pKa = 4.25DD59 pKa = 2.86AHH61 pKa = 7.17SLRR64 pKa = 11.84TPDD67 pKa = 3.59GAPAEE72 pKa = 4.26AVSDD76 pKa = 3.77EE77 pKa = 4.06FVTYY81 pKa = 10.12GAEE84 pKa = 4.21DD85 pKa = 3.96YY86 pKa = 11.25LEE88 pKa = 4.77KK89 pKa = 10.76SDD91 pKa = 6.3DD92 pKa = 3.73EE93 pKa = 4.54LLVAFEE99 pKa = 4.48TMVKK103 pKa = 9.87PMRR106 pKa = 11.84IGQLWCPAFNKK117 pKa = 10.21CSFISSIAMARR128 pKa = 11.84ALLLAPRR135 pKa = 11.84TSHH138 pKa = 6.16RR139 pKa = 11.84TMKK142 pKa = 10.54CFEE145 pKa = 4.89DD146 pKa = 4.26LVAAIYY152 pKa = 8.03TKK154 pKa = 10.88SDD156 pKa = 3.35FYY158 pKa = 11.69YY159 pKa = 10.59SEE161 pKa = 4.06EE162 pKa = 4.47CEE164 pKa = 4.84ADD166 pKa = 3.5DD167 pKa = 4.4AQIDD171 pKa = 3.63ISSRR175 pKa = 11.84DD176 pKa = 3.34VPGYY180 pKa = 10.54SFEE183 pKa = 4.16PWSRR187 pKa = 11.84TSGFEE192 pKa = 3.82PPPICEE198 pKa = 4.13ACDD201 pKa = 3.36MIMYY205 pKa = 8.23QCPCFDD211 pKa = 4.17FNALKK216 pKa = 10.42KK217 pKa = 10.37SCAEE221 pKa = 3.67RR222 pKa = 11.84TFADD226 pKa = 3.98DD227 pKa = 3.94YY228 pKa = 11.49VIEE231 pKa = 4.53GLDD234 pKa = 3.48GVVDD238 pKa = 3.83NATLLSNLGPFLVPVKK254 pKa = 10.21CQYY257 pKa = 9.26EE258 pKa = 4.27KK259 pKa = 11.27CPTPTIAIPPDD270 pKa = 3.47LNRR273 pKa = 11.84ATDD276 pKa = 3.45RR277 pKa = 11.84VDD279 pKa = 2.99INLVQSICDD288 pKa = 3.54STLPTHH294 pKa = 6.86SNYY297 pKa = 10.5DD298 pKa = 3.51DD299 pKa = 4.74SFHH302 pKa = 6.52QVFVEE307 pKa = 4.17SADD310 pKa = 3.56YY311 pKa = 11.12SIDD314 pKa = 3.74LDD316 pKa = 3.81HH317 pKa = 7.21VRR319 pKa = 11.84LRR321 pKa = 11.84QSDD324 pKa = 5.26LIAKK328 pKa = 9.64IPDD331 pKa = 3.93SGHH334 pKa = 6.42MIPVLNTGSGHH345 pKa = 5.95KK346 pKa = 10.01RR347 pKa = 11.84VGTTKK352 pKa = 10.48EE353 pKa = 3.68VLTAIKK359 pKa = 9.97KK360 pKa = 10.04RR361 pKa = 11.84NADD364 pKa = 3.49VPEE367 pKa = 5.43LGDD370 pKa = 3.75SVNLSRR376 pKa = 11.84LSKK379 pKa = 10.61AVAEE383 pKa = 4.47RR384 pKa = 11.84FFISYY389 pKa = 10.67INGNSLASSNFVNVVSNFHH408 pKa = 7.53DD409 pKa = 4.25YY410 pKa = 9.42MEE412 pKa = 4.41KK413 pKa = 9.79WKK415 pKa = 11.04SSGLSYY421 pKa = 11.44DD422 pKa = 4.55DD423 pKa = 5.94LPDD426 pKa = 3.54LHH428 pKa = 8.26AEE430 pKa = 3.82NLQFYY435 pKa = 10.96DD436 pKa = 4.19HH437 pKa = 6.9MIKK440 pKa = 10.37SDD442 pKa = 3.66VKK444 pKa = 10.5PVVSDD449 pKa = 3.14TLNIDD454 pKa = 3.49RR455 pKa = 11.84PVPATITYY463 pKa = 8.38HH464 pKa = 6.23KK465 pKa = 10.59KK466 pKa = 10.61SITSQFSPLFTALFEE481 pKa = 4.23RR482 pKa = 11.84FQRR485 pKa = 11.84CLRR488 pKa = 11.84EE489 pKa = 4.2RR490 pKa = 11.84IILPVGKK497 pKa = 10.03ISSLEE502 pKa = 3.74MAGFDD507 pKa = 4.72VKK509 pKa = 10.94NKK511 pKa = 9.77HH512 pKa = 6.23CLEE515 pKa = 4.69IDD517 pKa = 3.33LSKK520 pKa = 10.66FDD522 pKa = 3.64KK523 pKa = 11.1SQGEE527 pKa = 3.95FHH529 pKa = 7.12LLIQEE534 pKa = 4.92HH535 pKa = 6.57ILNGLGCPAPITKK548 pKa = 8.63WWCDD552 pKa = 3.85FHH554 pKa = 8.06RR555 pKa = 11.84FSYY558 pKa = 10.5IRR560 pKa = 11.84DD561 pKa = 3.2RR562 pKa = 11.84RR563 pKa = 11.84AGVGMPISFQRR574 pKa = 11.84RR575 pKa = 11.84TGDD578 pKa = 3.01ALTYY582 pKa = 10.06FGNTIVTMAEE592 pKa = 4.01FAWCYY597 pKa = 10.05DD598 pKa = 2.99TDD600 pKa = 3.72QFEE603 pKa = 4.62KK604 pKa = 11.13LLFSGDD610 pKa = 3.73DD611 pKa = 3.4SLGFSLLPPVGDD623 pKa = 3.38PSKK626 pKa = 10.39FTTLFNMEE634 pKa = 4.46AKK636 pKa = 10.42VMEE639 pKa = 4.39PAVPYY644 pKa = 10.13ICSKK648 pKa = 10.72FLLSDD653 pKa = 3.29EE654 pKa = 4.73FGNTFSVPDD663 pKa = 4.32PLRR666 pKa = 11.84EE667 pKa = 3.99VQRR670 pKa = 11.84LGTKK674 pKa = 10.07KK675 pKa = 10.14IPYY678 pKa = 9.12SDD680 pKa = 3.31NDD682 pKa = 3.45EE683 pKa = 4.15FLFAHH688 pKa = 6.5FMSFVDD694 pKa = 3.6RR695 pKa = 11.84LKK697 pKa = 11.07FLDD700 pKa = 5.08RR701 pKa = 11.84MSQSCIDD708 pKa = 3.38QLSIFFEE715 pKa = 4.35LKK717 pKa = 9.19YY718 pKa = 10.58KK719 pKa = 10.79KK720 pKa = 10.32SGEE723 pKa = 4.03EE724 pKa = 3.53AALMLGAFKK733 pKa = 10.76KK734 pKa = 9.49YY735 pKa = 7.71TANFQSYY742 pKa = 9.82KK743 pKa = 9.9EE744 pKa = 4.4LYY746 pKa = 9.56YY747 pKa = 10.53SDD749 pKa = 5.0RR750 pKa = 11.84RR751 pKa = 11.84QCEE754 pKa = 4.51LINSFCSTEE763 pKa = 3.83FRR765 pKa = 11.84VEE767 pKa = 3.76RR768 pKa = 11.84VNSNKK773 pKa = 8.44QRR775 pKa = 11.84KK776 pKa = 8.62NYY778 pKa = 10.18GIEE781 pKa = 3.73RR782 pKa = 11.84RR783 pKa = 11.84CNDD786 pKa = 2.98KK787 pKa = 10.85RR788 pKa = 11.84RR789 pKa = 11.84TPTGSYY795 pKa = 10.66GGGEE799 pKa = 3.92EE800 pKa = 5.26AEE802 pKa = 4.51TKK804 pKa = 10.38VSQTEE809 pKa = 4.25STGTRR814 pKa = 11.84SQKK817 pKa = 8.05SQRR820 pKa = 11.84EE821 pKa = 4.28SAFKK825 pKa = 10.74SQTIPLPTVLSSGWFGTDD843 pKa = 2.14RR844 pKa = 11.84VMPPCEE850 pKa = 3.87RR851 pKa = 11.84GGVTRR856 pKa = 11.84VV857 pKa = 2.88
MM1 pKa = 7.78AFPAPAFSLANLLNGSYY18 pKa = 10.84GVDD21 pKa = 3.06TPEE24 pKa = 3.93DD25 pKa = 3.67VEE27 pKa = 4.3RR28 pKa = 11.84LRR30 pKa = 11.84SEE32 pKa = 4.02QRR34 pKa = 11.84EE35 pKa = 4.09EE36 pKa = 3.47AAAACRR42 pKa = 11.84NYY44 pKa = 10.59RR45 pKa = 11.84PLPAVDD51 pKa = 3.51VSEE54 pKa = 4.73SVTEE58 pKa = 4.25DD59 pKa = 2.86AHH61 pKa = 7.17SLRR64 pKa = 11.84TPDD67 pKa = 3.59GAPAEE72 pKa = 4.26AVSDD76 pKa = 3.77EE77 pKa = 4.06FVTYY81 pKa = 10.12GAEE84 pKa = 4.21DD85 pKa = 3.96YY86 pKa = 11.25LEE88 pKa = 4.77KK89 pKa = 10.76SDD91 pKa = 6.3DD92 pKa = 3.73EE93 pKa = 4.54LLVAFEE99 pKa = 4.48TMVKK103 pKa = 9.87PMRR106 pKa = 11.84IGQLWCPAFNKK117 pKa = 10.21CSFISSIAMARR128 pKa = 11.84ALLLAPRR135 pKa = 11.84TSHH138 pKa = 6.16RR139 pKa = 11.84TMKK142 pKa = 10.54CFEE145 pKa = 4.89DD146 pKa = 4.26LVAAIYY152 pKa = 8.03TKK154 pKa = 10.88SDD156 pKa = 3.35FYY158 pKa = 11.69YY159 pKa = 10.59SEE161 pKa = 4.06EE162 pKa = 4.47CEE164 pKa = 4.84ADD166 pKa = 3.5DD167 pKa = 4.4AQIDD171 pKa = 3.63ISSRR175 pKa = 11.84DD176 pKa = 3.34VPGYY180 pKa = 10.54SFEE183 pKa = 4.16PWSRR187 pKa = 11.84TSGFEE192 pKa = 3.82PPPICEE198 pKa = 4.13ACDD201 pKa = 3.36MIMYY205 pKa = 8.23QCPCFDD211 pKa = 4.17FNALKK216 pKa = 10.42KK217 pKa = 10.37SCAEE221 pKa = 3.67RR222 pKa = 11.84TFADD226 pKa = 3.98DD227 pKa = 3.94YY228 pKa = 11.49VIEE231 pKa = 4.53GLDD234 pKa = 3.48GVVDD238 pKa = 3.83NATLLSNLGPFLVPVKK254 pKa = 10.21CQYY257 pKa = 9.26EE258 pKa = 4.27KK259 pKa = 11.27CPTPTIAIPPDD270 pKa = 3.47LNRR273 pKa = 11.84ATDD276 pKa = 3.45RR277 pKa = 11.84VDD279 pKa = 2.99INLVQSICDD288 pKa = 3.54STLPTHH294 pKa = 6.86SNYY297 pKa = 10.5DD298 pKa = 3.51DD299 pKa = 4.74SFHH302 pKa = 6.52QVFVEE307 pKa = 4.17SADD310 pKa = 3.56YY311 pKa = 11.12SIDD314 pKa = 3.74LDD316 pKa = 3.81HH317 pKa = 7.21VRR319 pKa = 11.84LRR321 pKa = 11.84QSDD324 pKa = 5.26LIAKK328 pKa = 9.64IPDD331 pKa = 3.93SGHH334 pKa = 6.42MIPVLNTGSGHH345 pKa = 5.95KK346 pKa = 10.01RR347 pKa = 11.84VGTTKK352 pKa = 10.48EE353 pKa = 3.68VLTAIKK359 pKa = 9.97KK360 pKa = 10.04RR361 pKa = 11.84NADD364 pKa = 3.49VPEE367 pKa = 5.43LGDD370 pKa = 3.75SVNLSRR376 pKa = 11.84LSKK379 pKa = 10.61AVAEE383 pKa = 4.47RR384 pKa = 11.84FFISYY389 pKa = 10.67INGNSLASSNFVNVVSNFHH408 pKa = 7.53DD409 pKa = 4.25YY410 pKa = 9.42MEE412 pKa = 4.41KK413 pKa = 9.79WKK415 pKa = 11.04SSGLSYY421 pKa = 11.44DD422 pKa = 4.55DD423 pKa = 5.94LPDD426 pKa = 3.54LHH428 pKa = 8.26AEE430 pKa = 3.82NLQFYY435 pKa = 10.96DD436 pKa = 4.19HH437 pKa = 6.9MIKK440 pKa = 10.37SDD442 pKa = 3.66VKK444 pKa = 10.5PVVSDD449 pKa = 3.14TLNIDD454 pKa = 3.49RR455 pKa = 11.84PVPATITYY463 pKa = 8.38HH464 pKa = 6.23KK465 pKa = 10.59KK466 pKa = 10.61SITSQFSPLFTALFEE481 pKa = 4.23RR482 pKa = 11.84FQRR485 pKa = 11.84CLRR488 pKa = 11.84EE489 pKa = 4.2RR490 pKa = 11.84IILPVGKK497 pKa = 10.03ISSLEE502 pKa = 3.74MAGFDD507 pKa = 4.72VKK509 pKa = 10.94NKK511 pKa = 9.77HH512 pKa = 6.23CLEE515 pKa = 4.69IDD517 pKa = 3.33LSKK520 pKa = 10.66FDD522 pKa = 3.64KK523 pKa = 11.1SQGEE527 pKa = 3.95FHH529 pKa = 7.12LLIQEE534 pKa = 4.92HH535 pKa = 6.57ILNGLGCPAPITKK548 pKa = 8.63WWCDD552 pKa = 3.85FHH554 pKa = 8.06RR555 pKa = 11.84FSYY558 pKa = 10.5IRR560 pKa = 11.84DD561 pKa = 3.2RR562 pKa = 11.84RR563 pKa = 11.84AGVGMPISFQRR574 pKa = 11.84RR575 pKa = 11.84TGDD578 pKa = 3.01ALTYY582 pKa = 10.06FGNTIVTMAEE592 pKa = 4.01FAWCYY597 pKa = 10.05DD598 pKa = 2.99TDD600 pKa = 3.72QFEE603 pKa = 4.62KK604 pKa = 11.13LLFSGDD610 pKa = 3.73DD611 pKa = 3.4SLGFSLLPPVGDD623 pKa = 3.38PSKK626 pKa = 10.39FTTLFNMEE634 pKa = 4.46AKK636 pKa = 10.42VMEE639 pKa = 4.39PAVPYY644 pKa = 10.13ICSKK648 pKa = 10.72FLLSDD653 pKa = 3.29EE654 pKa = 4.73FGNTFSVPDD663 pKa = 4.32PLRR666 pKa = 11.84EE667 pKa = 3.99VQRR670 pKa = 11.84LGTKK674 pKa = 10.07KK675 pKa = 10.14IPYY678 pKa = 9.12SDD680 pKa = 3.31NDD682 pKa = 3.45EE683 pKa = 4.15FLFAHH688 pKa = 6.5FMSFVDD694 pKa = 3.6RR695 pKa = 11.84LKK697 pKa = 11.07FLDD700 pKa = 5.08RR701 pKa = 11.84MSQSCIDD708 pKa = 3.38QLSIFFEE715 pKa = 4.35LKK717 pKa = 9.19YY718 pKa = 10.58KK719 pKa = 10.79KK720 pKa = 10.32SGEE723 pKa = 4.03EE724 pKa = 3.53AALMLGAFKK733 pKa = 10.76KK734 pKa = 9.49YY735 pKa = 7.71TANFQSYY742 pKa = 9.82KK743 pKa = 9.9EE744 pKa = 4.4LYY746 pKa = 9.56YY747 pKa = 10.53SDD749 pKa = 5.0RR750 pKa = 11.84RR751 pKa = 11.84QCEE754 pKa = 4.51LINSFCSTEE763 pKa = 3.83FRR765 pKa = 11.84VEE767 pKa = 3.76RR768 pKa = 11.84VNSNKK773 pKa = 8.44QRR775 pKa = 11.84KK776 pKa = 8.62NYY778 pKa = 10.18GIEE781 pKa = 3.73RR782 pKa = 11.84RR783 pKa = 11.84CNDD786 pKa = 2.98KK787 pKa = 10.85RR788 pKa = 11.84RR789 pKa = 11.84TPTGSYY795 pKa = 10.66GGGEE799 pKa = 3.92EE800 pKa = 5.26AEE802 pKa = 4.51TKK804 pKa = 10.38VSQTEE809 pKa = 4.25STGTRR814 pKa = 11.84SQKK817 pKa = 8.05SQRR820 pKa = 11.84EE821 pKa = 4.28SAFKK825 pKa = 10.74SQTIPLPTVLSSGWFGTDD843 pKa = 2.14RR844 pKa = 11.84VMPPCEE850 pKa = 3.87RR851 pKa = 11.84GGVTRR856 pKa = 11.84VV857 pKa = 2.88
Molecular weight: 96.71 kDa
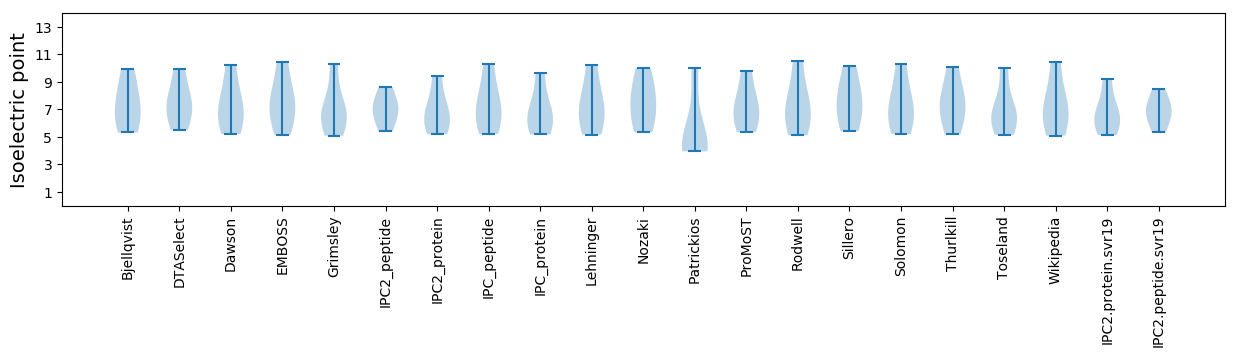
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q00271|MVP_CMVFN Movement protein OS=Cucumber mosaic virus (strain FNY) OX=12307 GN=ORF3a PE=3 SV=1
MM1 pKa = 7.98DD2 pKa = 4.94KK3 pKa = 11.01SEE5 pKa = 4.19STSAGRR11 pKa = 11.84NRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84PRR18 pKa = 11.84RR19 pKa = 11.84GSRR22 pKa = 11.84SAPSSADD29 pKa = 2.97ANFRR33 pKa = 11.84VLSQQLSRR41 pKa = 11.84LNKK44 pKa = 8.14TLAAGRR50 pKa = 11.84PTINHH55 pKa = 5.52PTFVGSEE62 pKa = 4.04RR63 pKa = 11.84CRR65 pKa = 11.84PGYY68 pKa = 9.09TFTSITLKK76 pKa = 10.38PPKK79 pKa = 9.04IDD81 pKa = 3.14RR82 pKa = 11.84GSYY85 pKa = 7.96YY86 pKa = 10.78GKK88 pKa = 10.51RR89 pKa = 11.84LLLPDD94 pKa = 4.13SVTEE98 pKa = 3.94YY99 pKa = 11.07DD100 pKa = 3.37KK101 pKa = 11.59KK102 pKa = 10.76LVSRR106 pKa = 11.84IQIRR110 pKa = 11.84VNPLPKK116 pKa = 9.66FDD118 pKa = 3.58STVWVTVRR126 pKa = 11.84KK127 pKa = 10.07VPASSDD133 pKa = 3.39LSVAAISAMFADD145 pKa = 5.23GASPVLVYY153 pKa = 9.71QYY155 pKa = 11.09AASGVQANNKK165 pKa = 9.21LLYY168 pKa = 10.24DD169 pKa = 4.23LSAMRR174 pKa = 11.84ADD176 pKa = 3.22IGDD179 pKa = 3.54MRR181 pKa = 11.84KK182 pKa = 9.47YY183 pKa = 10.88AVLVYY188 pKa = 10.8SKK190 pKa = 11.21DD191 pKa = 3.59DD192 pKa = 3.71ALEE195 pKa = 4.01TDD197 pKa = 3.9EE198 pKa = 6.0LVLHH202 pKa = 6.52VDD204 pKa = 3.74IEE206 pKa = 4.39HH207 pKa = 6.04QRR209 pKa = 11.84IPTSGVLPVV218 pKa = 3.49
MM1 pKa = 7.98DD2 pKa = 4.94KK3 pKa = 11.01SEE5 pKa = 4.19STSAGRR11 pKa = 11.84NRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84PRR18 pKa = 11.84RR19 pKa = 11.84GSRR22 pKa = 11.84SAPSSADD29 pKa = 2.97ANFRR33 pKa = 11.84VLSQQLSRR41 pKa = 11.84LNKK44 pKa = 8.14TLAAGRR50 pKa = 11.84PTINHH55 pKa = 5.52PTFVGSEE62 pKa = 4.04RR63 pKa = 11.84CRR65 pKa = 11.84PGYY68 pKa = 9.09TFTSITLKK76 pKa = 10.38PPKK79 pKa = 9.04IDD81 pKa = 3.14RR82 pKa = 11.84GSYY85 pKa = 7.96YY86 pKa = 10.78GKK88 pKa = 10.51RR89 pKa = 11.84LLLPDD94 pKa = 4.13SVTEE98 pKa = 3.94YY99 pKa = 11.07DD100 pKa = 3.37KK101 pKa = 11.59KK102 pKa = 10.76LVSRR106 pKa = 11.84IQIRR110 pKa = 11.84VNPLPKK116 pKa = 9.66FDD118 pKa = 3.58STVWVTVRR126 pKa = 11.84KK127 pKa = 10.07VPASSDD133 pKa = 3.39LSVAAISAMFADD145 pKa = 5.23GASPVLVYY153 pKa = 9.71QYY155 pKa = 11.09AASGVQANNKK165 pKa = 9.21LLYY168 pKa = 10.24DD169 pKa = 4.23LSAMRR174 pKa = 11.84ADD176 pKa = 3.22IGDD179 pKa = 3.54MRR181 pKa = 11.84KK182 pKa = 9.47YY183 pKa = 10.88AVLVYY188 pKa = 10.8SKK190 pKa = 11.21DD191 pKa = 3.59DD192 pKa = 3.71ALEE195 pKa = 4.01TDD197 pKa = 3.9EE198 pKa = 6.0LVLHH202 pKa = 6.52VDD204 pKa = 3.74IEE206 pKa = 4.39HH207 pKa = 6.04QRR209 pKa = 11.84IPTSGVLPVV218 pKa = 3.49
Molecular weight: 24.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2457 |
110 |
993 |
491.4 |
55.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.367 ± 0.465 | 2.401 ± 0.323 |
6.349 ± 0.489 | 5.983 ± 0.618 |
4.843 ± 0.7 | 5.413 ± 0.209 |
2.239 ± 0.222 | 4.762 ± 0.468 |
5.454 ± 0.266 | 8.262 ± 0.185 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.605 ± 0.316 | 3.663 ± 0.298 |
4.477 ± 0.674 | 3.378 ± 0.257 |
6.431 ± 0.671 | 9.28 ± 0.391 |
6.064 ± 0.499 | 6.919 ± 0.492 |
1.018 ± 0.153 | 3.093 ± 0.26 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
