
Circoviridae 7 LDMD-2013
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; environmental samples
Average proteome isoelectric point is 7.62
Get precalculated fractions of proteins
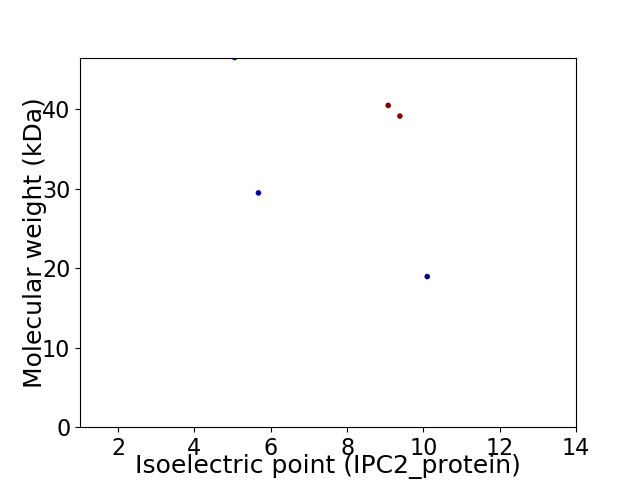
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|S5TNC7|S5TNC7_9CIRC Replication-associated protein OS=Circoviridae 7 LDMD-2013 OX=1379711 PE=4 SV=1
MM1 pKa = 7.84RR2 pKa = 11.84GNPGEE7 pKa = 4.28ASGEE11 pKa = 3.93ALDD14 pKa = 5.13DD15 pKa = 3.79VRR17 pKa = 11.84TLAVLGNWDD26 pKa = 3.55TTKK29 pKa = 11.12ANPAVTNYY37 pKa = 9.58PLSASNTISRR47 pKa = 11.84RR48 pKa = 11.84YY49 pKa = 9.87NGARR53 pKa = 11.84QQNWLNVEE61 pKa = 4.18PLSEE65 pKa = 5.08PGTSLYY71 pKa = 10.46GAPLPEE77 pKa = 4.27QYY79 pKa = 10.59DD80 pKa = 3.8VQVGKK85 pKa = 10.82GYY87 pKa = 10.32IEE89 pKa = 3.7FTVKK93 pKa = 10.87NEE95 pKa = 3.89GTRR98 pKa = 11.84QAVVEE103 pKa = 4.14LVLFKK108 pKa = 10.76PNKK111 pKa = 9.0NAFNKK116 pKa = 10.2PYY118 pKa = 10.28LIDD121 pKa = 3.87EE122 pKa = 4.74APSAEE127 pKa = 4.03IAPNIAEE134 pKa = 4.34NVWYY138 pKa = 9.17WLNRR142 pKa = 11.84QNGEE146 pKa = 4.07QYY148 pKa = 9.96VQNIMSTRR156 pKa = 11.84ARR158 pKa = 11.84ALAKK162 pKa = 10.16RR163 pKa = 11.84VGPTDD168 pKa = 3.36TNVPIDD174 pKa = 4.32KK175 pKa = 10.04PDD177 pKa = 3.73SNDD180 pKa = 3.14ILISPYY186 pKa = 11.11VNFLPEE192 pKa = 3.98SSFKK196 pKa = 11.08NVVGNSQAFKK206 pKa = 10.5RR207 pKa = 11.84PWMLDD212 pKa = 2.87GAGNGNVIVQSTVAEE227 pKa = 4.1EE228 pKa = 4.2MAASTSNIAEE238 pKa = 4.25VEE240 pKa = 4.05GGEE243 pKa = 4.58LNDD246 pKa = 4.15GEE248 pKa = 5.04AXGRR252 pKa = 11.84WKK254 pKa = 10.88NPYY257 pKa = 8.93TSIGRR262 pKa = 11.84AFAAVPAQGKK272 pKa = 7.47RR273 pKa = 11.84TIRR276 pKa = 11.84IPLPKK281 pKa = 9.92LRR283 pKa = 11.84YY284 pKa = 9.27NPSTSSDD291 pKa = 3.51YY292 pKa = 11.7SKK294 pKa = 11.34LDD296 pKa = 3.21DD297 pKa = 5.01DD298 pKa = 4.67FAIRR302 pKa = 11.84PQSGTSATLYY312 pKa = 7.03PTIMNPNSIMVAMSLNGARR331 pKa = 11.84SDD333 pKa = 3.79FFEE336 pKa = 4.75TKK338 pKa = 10.4DD339 pKa = 3.43SEE341 pKa = 4.51AKK343 pKa = 10.39FRR345 pKa = 11.84GQDD348 pKa = 3.12FTAAKK353 pKa = 9.82VYY355 pKa = 10.78VDD357 pKa = 3.74AVYY360 pKa = 10.86KK361 pKa = 8.92EE362 pKa = 4.45TVYY365 pKa = 10.62PCAMDD370 pKa = 3.55TQEE373 pKa = 4.64TKK375 pKa = 9.78TAFNFGTPYY384 pKa = 7.97FTNMGEE390 pKa = 4.29VPVGSSYY397 pKa = 10.56WPGVVQDD404 pKa = 4.24AANVQNTVSFGTSIGAGVVQTPGTT428 pKa = 3.63
MM1 pKa = 7.84RR2 pKa = 11.84GNPGEE7 pKa = 4.28ASGEE11 pKa = 3.93ALDD14 pKa = 5.13DD15 pKa = 3.79VRR17 pKa = 11.84TLAVLGNWDD26 pKa = 3.55TTKK29 pKa = 11.12ANPAVTNYY37 pKa = 9.58PLSASNTISRR47 pKa = 11.84RR48 pKa = 11.84YY49 pKa = 9.87NGARR53 pKa = 11.84QQNWLNVEE61 pKa = 4.18PLSEE65 pKa = 5.08PGTSLYY71 pKa = 10.46GAPLPEE77 pKa = 4.27QYY79 pKa = 10.59DD80 pKa = 3.8VQVGKK85 pKa = 10.82GYY87 pKa = 10.32IEE89 pKa = 3.7FTVKK93 pKa = 10.87NEE95 pKa = 3.89GTRR98 pKa = 11.84QAVVEE103 pKa = 4.14LVLFKK108 pKa = 10.76PNKK111 pKa = 9.0NAFNKK116 pKa = 10.2PYY118 pKa = 10.28LIDD121 pKa = 3.87EE122 pKa = 4.74APSAEE127 pKa = 4.03IAPNIAEE134 pKa = 4.34NVWYY138 pKa = 9.17WLNRR142 pKa = 11.84QNGEE146 pKa = 4.07QYY148 pKa = 9.96VQNIMSTRR156 pKa = 11.84ARR158 pKa = 11.84ALAKK162 pKa = 10.16RR163 pKa = 11.84VGPTDD168 pKa = 3.36TNVPIDD174 pKa = 4.32KK175 pKa = 10.04PDD177 pKa = 3.73SNDD180 pKa = 3.14ILISPYY186 pKa = 11.11VNFLPEE192 pKa = 3.98SSFKK196 pKa = 11.08NVVGNSQAFKK206 pKa = 10.5RR207 pKa = 11.84PWMLDD212 pKa = 2.87GAGNGNVIVQSTVAEE227 pKa = 4.1EE228 pKa = 4.2MAASTSNIAEE238 pKa = 4.25VEE240 pKa = 4.05GGEE243 pKa = 4.58LNDD246 pKa = 4.15GEE248 pKa = 5.04AXGRR252 pKa = 11.84WKK254 pKa = 10.88NPYY257 pKa = 8.93TSIGRR262 pKa = 11.84AFAAVPAQGKK272 pKa = 7.47RR273 pKa = 11.84TIRR276 pKa = 11.84IPLPKK281 pKa = 9.92LRR283 pKa = 11.84YY284 pKa = 9.27NPSTSSDD291 pKa = 3.51YY292 pKa = 11.7SKK294 pKa = 11.34LDD296 pKa = 3.21DD297 pKa = 5.01DD298 pKa = 4.67FAIRR302 pKa = 11.84PQSGTSATLYY312 pKa = 7.03PTIMNPNSIMVAMSLNGARR331 pKa = 11.84SDD333 pKa = 3.79FFEE336 pKa = 4.75TKK338 pKa = 10.4DD339 pKa = 3.43SEE341 pKa = 4.51AKK343 pKa = 10.39FRR345 pKa = 11.84GQDD348 pKa = 3.12FTAAKK353 pKa = 9.82VYY355 pKa = 10.78VDD357 pKa = 3.74AVYY360 pKa = 10.86KK361 pKa = 8.92EE362 pKa = 4.45TVYY365 pKa = 10.62PCAMDD370 pKa = 3.55TQEE373 pKa = 4.64TKK375 pKa = 9.78TAFNFGTPYY384 pKa = 7.97FTNMGEE390 pKa = 4.29VPVGSSYY397 pKa = 10.56WPGVVQDD404 pKa = 4.24AANVQNTVSFGTSIGAGVVQTPGTT428 pKa = 3.63
Molecular weight: 46.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5TMV3|S5TMV3_9CIRC Uncharacterized protein OS=Circoviridae 7 LDMD-2013 OX=1379711 PE=4 SV=1
MM1 pKa = 7.66GKK3 pKa = 9.73NDD5 pKa = 3.91TYY7 pKa = 10.69ICSKK11 pKa = 10.14KK12 pKa = 10.27RR13 pKa = 11.84CTFYY17 pKa = 10.61STILLTYY24 pKa = 8.58WSRR27 pKa = 11.84ARR29 pKa = 11.84MAWTRR34 pKa = 11.84GRR36 pKa = 11.84TRR38 pKa = 11.84SGAYY42 pKa = 9.04YY43 pKa = 9.98QAAEE47 pKa = 4.24QIADD51 pKa = 3.55EE52 pKa = 4.21YY53 pKa = 10.63LRR55 pKa = 11.84RR56 pKa = 11.84VGIQDD61 pKa = 2.73RR62 pKa = 11.84RR63 pKa = 11.84MRR65 pKa = 11.84QVAIAGITAGLQRR78 pKa = 11.84GVTNFRR84 pKa = 11.84GLIPYY89 pKa = 9.39IRR91 pKa = 11.84EE92 pKa = 4.19SLPSLPVVDD101 pKa = 4.69PLFMARR107 pKa = 11.84SFRR110 pKa = 11.84DD111 pKa = 3.17AMVSITGEE119 pKa = 3.94VTRR122 pKa = 11.84PSLSAPAPSTSSSVGAQGSNMTSPQTQTRR151 pKa = 11.84MLLRR155 pKa = 11.84RR156 pKa = 11.84PAPGSSTKK164 pKa = 7.98TVKK167 pKa = 9.89RR168 pKa = 11.84HH169 pKa = 4.83KK170 pKa = 10.56KK171 pKa = 8.29KK172 pKa = 11.23GNMSGLDD179 pKa = 3.6PNTVQTYY186 pKa = 9.21PRR188 pKa = 11.84LTVSMPTDD196 pKa = 3.21KK197 pKa = 10.8TLLRR201 pKa = 11.84SSFAQFKK208 pKa = 10.25KK209 pKa = 9.44WFDD212 pKa = 3.27GHH214 pKa = 6.7RR215 pKa = 11.84VLTNSFSFIMEE226 pKa = 3.92STQGKK231 pKa = 9.63RR232 pKa = 11.84GNLLIPLRR240 pKa = 11.84CDD242 pKa = 2.92PCIYY246 pKa = 10.46GGRR249 pKa = 11.84THH251 pKa = 6.52EE252 pKa = 5.27QDD254 pKa = 3.17TADD257 pKa = 3.08NYY259 pKa = 10.85YY260 pKa = 10.75RR261 pKa = 11.84FVTGASWGQDD271 pKa = 3.1VNLPDD276 pKa = 4.17PVDD279 pKa = 3.53GKK281 pKa = 10.74KK282 pKa = 10.19VQDD285 pKa = 3.46IGPAGHH291 pKa = 5.79YY292 pKa = 7.83QHH294 pKa = 6.8VVGDD298 pKa = 3.95LMRR301 pKa = 11.84TGYY304 pKa = 10.53SDD306 pKa = 3.45STVCEE311 pKa = 4.36PVTKK315 pKa = 10.6DD316 pKa = 3.65LDD318 pKa = 3.78LSIPGNSITQQTATAKK334 pKa = 10.5HH335 pKa = 5.99MIVPGLTASSFRR347 pKa = 11.84AGIMAVEE354 pKa = 4.24
MM1 pKa = 7.66GKK3 pKa = 9.73NDD5 pKa = 3.91TYY7 pKa = 10.69ICSKK11 pKa = 10.14KK12 pKa = 10.27RR13 pKa = 11.84CTFYY17 pKa = 10.61STILLTYY24 pKa = 8.58WSRR27 pKa = 11.84ARR29 pKa = 11.84MAWTRR34 pKa = 11.84GRR36 pKa = 11.84TRR38 pKa = 11.84SGAYY42 pKa = 9.04YY43 pKa = 9.98QAAEE47 pKa = 4.24QIADD51 pKa = 3.55EE52 pKa = 4.21YY53 pKa = 10.63LRR55 pKa = 11.84RR56 pKa = 11.84VGIQDD61 pKa = 2.73RR62 pKa = 11.84RR63 pKa = 11.84MRR65 pKa = 11.84QVAIAGITAGLQRR78 pKa = 11.84GVTNFRR84 pKa = 11.84GLIPYY89 pKa = 9.39IRR91 pKa = 11.84EE92 pKa = 4.19SLPSLPVVDD101 pKa = 4.69PLFMARR107 pKa = 11.84SFRR110 pKa = 11.84DD111 pKa = 3.17AMVSITGEE119 pKa = 3.94VTRR122 pKa = 11.84PSLSAPAPSTSSSVGAQGSNMTSPQTQTRR151 pKa = 11.84MLLRR155 pKa = 11.84RR156 pKa = 11.84PAPGSSTKK164 pKa = 7.98TVKK167 pKa = 9.89RR168 pKa = 11.84HH169 pKa = 4.83KK170 pKa = 10.56KK171 pKa = 8.29KK172 pKa = 11.23GNMSGLDD179 pKa = 3.6PNTVQTYY186 pKa = 9.21PRR188 pKa = 11.84LTVSMPTDD196 pKa = 3.21KK197 pKa = 10.8TLLRR201 pKa = 11.84SSFAQFKK208 pKa = 10.25KK209 pKa = 9.44WFDD212 pKa = 3.27GHH214 pKa = 6.7RR215 pKa = 11.84VLTNSFSFIMEE226 pKa = 3.92STQGKK231 pKa = 9.63RR232 pKa = 11.84GNLLIPLRR240 pKa = 11.84CDD242 pKa = 2.92PCIYY246 pKa = 10.46GGRR249 pKa = 11.84THH251 pKa = 6.52EE252 pKa = 5.27QDD254 pKa = 3.17TADD257 pKa = 3.08NYY259 pKa = 10.85YY260 pKa = 10.75RR261 pKa = 11.84FVTGASWGQDD271 pKa = 3.1VNLPDD276 pKa = 4.17PVDD279 pKa = 3.53GKK281 pKa = 10.74KK282 pKa = 10.19VQDD285 pKa = 3.46IGPAGHH291 pKa = 5.79YY292 pKa = 7.83QHH294 pKa = 6.8VVGDD298 pKa = 3.95LMRR301 pKa = 11.84TGYY304 pKa = 10.53SDD306 pKa = 3.45STVCEE311 pKa = 4.36PVTKK315 pKa = 10.6DD316 pKa = 3.65LDD318 pKa = 3.78LSIPGNSITQQTATAKK334 pKa = 10.5HH335 pKa = 5.99MIVPGLTASSFRR347 pKa = 11.84AGIMAVEE354 pKa = 4.24
Molecular weight: 39.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1568 |
153 |
428 |
313.6 |
34.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.334 ± 0.772 | 1.339 ± 0.52 |
4.911 ± 0.542 | 5.102 ± 0.751 |
3.827 ± 0.212 | 6.888 ± 0.577 |
1.467 ± 0.583 | 4.273 ± 0.364 |
4.592 ± 0.665 | 7.079 ± 1.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.423 ± 0.506 | 5.293 ± 1.137 |
7.079 ± 0.329 | 3.763 ± 0.347 |
8.099 ± 1.803 | 7.079 ± 0.63 |
7.589 ± 0.647 | 6.378 ± 0.634 |
1.531 ± 0.129 | 3.444 ± 0.53 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
