
Melandrium yellow fleck virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Bromovirus
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
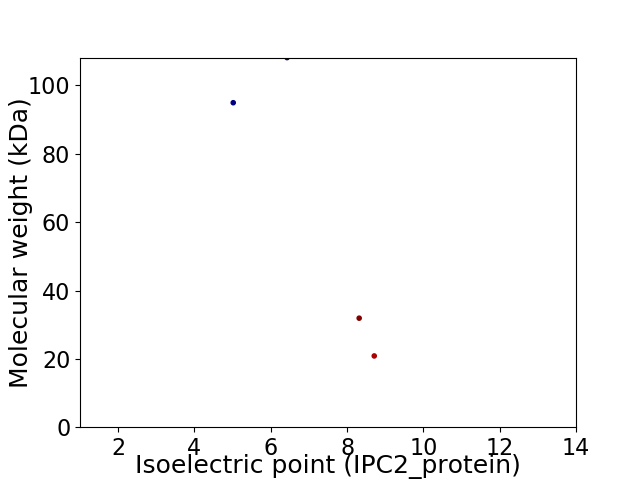
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
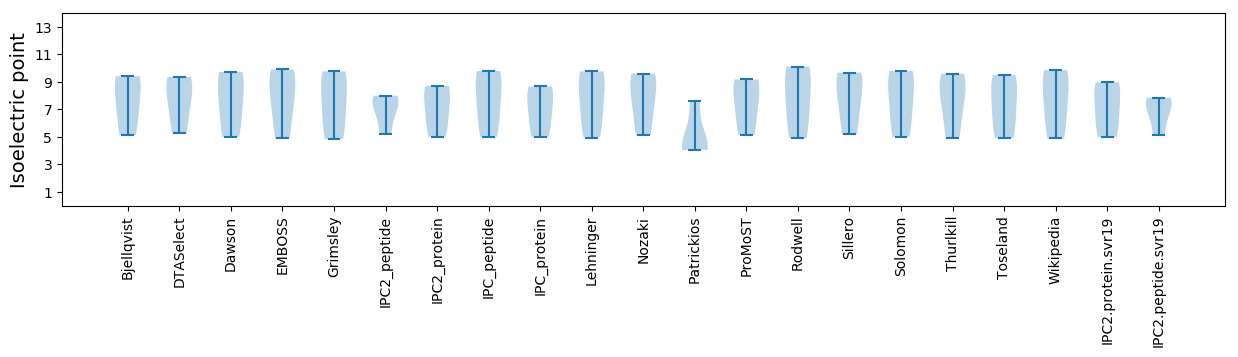
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C8KH77|C8KH77_9BROM Movement protein OS=Melandrium yellow fleck virus OX=545259 GN=3a PE=4 SV=1
MM1 pKa = 7.45AFEE4 pKa = 4.71IEE6 pKa = 4.35YY7 pKa = 9.88EE8 pKa = 4.39VPSFQWMLDD17 pKa = 3.17TSRR20 pKa = 11.84LDD22 pKa = 3.53NYY24 pKa = 10.83SLNNGEE30 pKa = 4.52PSNIIPVDD38 pKa = 3.71DD39 pKa = 4.85SIEE42 pKa = 4.03IVADD46 pKa = 3.91GTLSSYY52 pKa = 10.96LHH54 pKa = 6.13AVKK57 pKa = 9.92PLEE60 pKa = 4.22IGGSYY65 pKa = 10.0EE66 pKa = 3.95PPFEE70 pKa = 3.94QSRR73 pKa = 11.84WAQCCEE79 pKa = 3.44NAINVSKK86 pKa = 10.48MKK88 pKa = 10.73GVILKK93 pKa = 8.24PLPEE97 pKa = 4.15VARR100 pKa = 11.84ILYY103 pKa = 10.17LDD105 pKa = 4.18LDD107 pKa = 4.12EE108 pKa = 6.02PIVDD112 pKa = 3.86EE113 pKa = 5.59SEE115 pKa = 3.97VDD117 pKa = 3.09DD118 pKa = 4.35WRR120 pKa = 11.84PIDD123 pKa = 3.74TSDD126 pKa = 3.85GYY128 pKa = 11.32DD129 pKa = 3.76PEE131 pKa = 4.46TSDD134 pKa = 4.45WDD136 pKa = 3.51PTYY139 pKa = 11.29LDD141 pKa = 4.07RR142 pKa = 11.84EE143 pKa = 4.35ADD145 pKa = 3.35AKK147 pKa = 11.11AYY149 pKa = 10.21GHH151 pKa = 7.39LLPSYY156 pKa = 6.88QQKK159 pKa = 10.36EE160 pKa = 4.02IAVSDD165 pKa = 4.09VEE167 pKa = 4.48TDD169 pKa = 3.43RR170 pKa = 11.84QNQDD174 pKa = 2.7VKK176 pKa = 11.37NVLLNPDD183 pKa = 4.34PLPARR188 pKa = 11.84EE189 pKa = 4.03FTLGDD194 pKa = 2.71RR195 pKa = 11.84FMPIEE200 pKa = 4.2EE201 pKa = 4.33EE202 pKa = 4.16FGGTGADD209 pKa = 3.26YY210 pKa = 11.18DD211 pKa = 3.98VTLNLINPVEE221 pKa = 4.27SRR223 pKa = 11.84VARR226 pKa = 11.84VVDD229 pKa = 3.5TLGGLEE235 pKa = 4.18SGVTLTSPLFLEE247 pKa = 4.32RR248 pKa = 11.84VSLNRR253 pKa = 11.84VDD255 pKa = 5.05AVGHH259 pKa = 6.08HH260 pKa = 6.44MLPTHH265 pKa = 7.31AYY267 pKa = 10.21FDD269 pKa = 3.68DD270 pKa = 4.06TYY272 pKa = 11.36YY273 pKa = 10.79QALEE277 pKa = 4.13EE278 pKa = 4.21NADD281 pKa = 3.57YY282 pKa = 11.36SSDD285 pKa = 3.67FNKK288 pKa = 10.64LKK290 pKa = 10.45IKK292 pKa = 10.64QSDD295 pKa = 3.67VDD297 pKa = 3.6WYY299 pKa = 11.05RR300 pKa = 11.84DD301 pKa = 3.19PDD303 pKa = 3.73KK304 pKa = 11.26YY305 pKa = 11.27YY306 pKa = 10.84EE307 pKa = 4.43PNLTIGSFQKK317 pKa = 10.71RR318 pKa = 11.84IGTQKK323 pKa = 9.58TVLTALKK330 pKa = 9.92KK331 pKa = 10.5RR332 pKa = 11.84NADD335 pKa = 3.46VPEE338 pKa = 4.18MSDD341 pKa = 3.53AIDD344 pKa = 3.48VKK346 pKa = 10.68RR347 pKa = 11.84VARR350 pKa = 11.84EE351 pKa = 3.74VADD354 pKa = 3.98KK355 pKa = 10.91FISSYY360 pKa = 11.46LNVDD364 pKa = 3.6GKK366 pKa = 10.92DD367 pKa = 3.51CFINGLNVMAKK378 pKa = 10.47GLDD381 pKa = 3.9YY382 pKa = 10.6YY383 pKa = 11.2KK384 pKa = 10.33KK385 pKa = 9.38WKK387 pKa = 9.68DD388 pKa = 3.19HH389 pKa = 6.82RR390 pKa = 11.84EE391 pKa = 3.95LSGVTIATGEE401 pKa = 3.85NLQRR405 pKa = 11.84YY406 pKa = 6.22QHH408 pKa = 6.22MIKK411 pKa = 9.56TDD413 pKa = 3.31IKK415 pKa = 10.77PVVTDD420 pKa = 3.53TLHH423 pKa = 7.11LDD425 pKa = 3.35RR426 pKa = 11.84AIAATITFHH435 pKa = 6.66GKK437 pKa = 10.12GVTSCFSPFFTACFEE452 pKa = 4.22NFSMALKK459 pKa = 10.51SRR461 pKa = 11.84FVVPIGKK468 pKa = 9.37ISSLEE473 pKa = 3.86IPNVSLNNKK482 pKa = 8.32WFLEE486 pKa = 4.07ADD488 pKa = 3.42LSKK491 pKa = 10.63FDD493 pKa = 3.7KK494 pKa = 11.18SQGEE498 pKa = 3.98LHH500 pKa = 7.06LEE502 pKa = 4.06FQRR505 pKa = 11.84EE506 pKa = 3.85ILHH509 pKa = 6.78AIGFPVHH516 pKa = 6.79LSNWWADD523 pKa = 4.13FHH525 pKa = 7.55RR526 pKa = 11.84EE527 pKa = 4.21SYY529 pKa = 11.39LNDD532 pKa = 3.54PNAKK536 pKa = 9.64VSLPVSFQRR545 pKa = 11.84RR546 pKa = 11.84TGDD549 pKa = 2.78AFTYY553 pKa = 10.13FGNTIVTMAMMAYY566 pKa = 8.3TFDD569 pKa = 3.62MNLPEE574 pKa = 4.27LAIFSGDD581 pKa = 3.91DD582 pKa = 3.31SLLVLNDD589 pKa = 3.85KK590 pKa = 10.79PSIDD594 pKa = 3.27TDD596 pKa = 3.19IFQRR600 pKa = 11.84LFNMEE605 pKa = 4.22VKK607 pKa = 10.26IMEE610 pKa = 4.38PSVPYY615 pKa = 10.34VCSKK619 pKa = 10.75FLLEE623 pKa = 4.32TEE625 pKa = 4.41LGSVVSVPDD634 pKa = 4.06PLRR637 pKa = 11.84EE638 pKa = 3.71IQRR641 pKa = 11.84MSKK644 pKa = 10.36RR645 pKa = 11.84KK646 pKa = 8.98ILKK649 pKa = 10.14DD650 pKa = 3.49PEE652 pKa = 3.78VLRR655 pKa = 11.84AHH657 pKa = 6.16FTSFKK662 pKa = 10.89DD663 pKa = 3.25RR664 pKa = 11.84MRR666 pKa = 11.84FLDD669 pKa = 3.56VLDD672 pKa = 4.82DD673 pKa = 4.07RR674 pKa = 11.84MISILCRR681 pKa = 11.84YY682 pKa = 9.56VSLKK686 pKa = 9.64YY687 pKa = 10.43LKK689 pKa = 10.18PGLEE693 pKa = 3.72QDD695 pKa = 3.3VRR697 pKa = 11.84EE698 pKa = 4.21ALACFSYY705 pKa = 10.75YY706 pKa = 10.25SEE708 pKa = 4.31SFFRR712 pKa = 11.84FAEE715 pKa = 5.05LYY717 pKa = 9.82AQDD720 pKa = 3.87GKK722 pKa = 10.95FVYY725 pKa = 9.54QVKK728 pKa = 10.62DD729 pKa = 3.92PISHH733 pKa = 6.4HH734 pKa = 6.7RR735 pKa = 11.84GDD737 pKa = 4.53EE738 pKa = 3.99ILGSRR743 pKa = 11.84EE744 pKa = 3.38KK745 pKa = 11.16DD746 pKa = 2.6GDD748 pKa = 4.06YY749 pKa = 10.72FHH751 pKa = 7.06NWHH754 pKa = 6.49NPVFPRR760 pKa = 11.84ILDD763 pKa = 3.37KK764 pKa = 10.96AVRR767 pKa = 11.84VFGKK771 pKa = 10.68YY772 pKa = 8.11STDD775 pKa = 3.4YY776 pKa = 11.26SSEE779 pKa = 4.06NYY781 pKa = 9.16KK782 pKa = 10.31KK783 pKa = 10.79KK784 pKa = 10.84SMKK787 pKa = 10.5CEE789 pKa = 3.6EE790 pKa = 4.52DD791 pKa = 3.22KK792 pKa = 11.31LFRR795 pKa = 11.84KK796 pKa = 9.73SLSLAYY802 pKa = 9.87DD803 pKa = 3.24QRR805 pKa = 11.84DD806 pKa = 3.43LLKK809 pKa = 11.06NKK811 pKa = 9.74LKK813 pKa = 10.36HH814 pKa = 7.08DD815 pKa = 4.08RR816 pKa = 11.84LLDD819 pKa = 3.62EE820 pKa = 4.64TASSSFSKK828 pKa = 10.95
MM1 pKa = 7.45AFEE4 pKa = 4.71IEE6 pKa = 4.35YY7 pKa = 9.88EE8 pKa = 4.39VPSFQWMLDD17 pKa = 3.17TSRR20 pKa = 11.84LDD22 pKa = 3.53NYY24 pKa = 10.83SLNNGEE30 pKa = 4.52PSNIIPVDD38 pKa = 3.71DD39 pKa = 4.85SIEE42 pKa = 4.03IVADD46 pKa = 3.91GTLSSYY52 pKa = 10.96LHH54 pKa = 6.13AVKK57 pKa = 9.92PLEE60 pKa = 4.22IGGSYY65 pKa = 10.0EE66 pKa = 3.95PPFEE70 pKa = 3.94QSRR73 pKa = 11.84WAQCCEE79 pKa = 3.44NAINVSKK86 pKa = 10.48MKK88 pKa = 10.73GVILKK93 pKa = 8.24PLPEE97 pKa = 4.15VARR100 pKa = 11.84ILYY103 pKa = 10.17LDD105 pKa = 4.18LDD107 pKa = 4.12EE108 pKa = 6.02PIVDD112 pKa = 3.86EE113 pKa = 5.59SEE115 pKa = 3.97VDD117 pKa = 3.09DD118 pKa = 4.35WRR120 pKa = 11.84PIDD123 pKa = 3.74TSDD126 pKa = 3.85GYY128 pKa = 11.32DD129 pKa = 3.76PEE131 pKa = 4.46TSDD134 pKa = 4.45WDD136 pKa = 3.51PTYY139 pKa = 11.29LDD141 pKa = 4.07RR142 pKa = 11.84EE143 pKa = 4.35ADD145 pKa = 3.35AKK147 pKa = 11.11AYY149 pKa = 10.21GHH151 pKa = 7.39LLPSYY156 pKa = 6.88QQKK159 pKa = 10.36EE160 pKa = 4.02IAVSDD165 pKa = 4.09VEE167 pKa = 4.48TDD169 pKa = 3.43RR170 pKa = 11.84QNQDD174 pKa = 2.7VKK176 pKa = 11.37NVLLNPDD183 pKa = 4.34PLPARR188 pKa = 11.84EE189 pKa = 4.03FTLGDD194 pKa = 2.71RR195 pKa = 11.84FMPIEE200 pKa = 4.2EE201 pKa = 4.33EE202 pKa = 4.16FGGTGADD209 pKa = 3.26YY210 pKa = 11.18DD211 pKa = 3.98VTLNLINPVEE221 pKa = 4.27SRR223 pKa = 11.84VARR226 pKa = 11.84VVDD229 pKa = 3.5TLGGLEE235 pKa = 4.18SGVTLTSPLFLEE247 pKa = 4.32RR248 pKa = 11.84VSLNRR253 pKa = 11.84VDD255 pKa = 5.05AVGHH259 pKa = 6.08HH260 pKa = 6.44MLPTHH265 pKa = 7.31AYY267 pKa = 10.21FDD269 pKa = 3.68DD270 pKa = 4.06TYY272 pKa = 11.36YY273 pKa = 10.79QALEE277 pKa = 4.13EE278 pKa = 4.21NADD281 pKa = 3.57YY282 pKa = 11.36SSDD285 pKa = 3.67FNKK288 pKa = 10.64LKK290 pKa = 10.45IKK292 pKa = 10.64QSDD295 pKa = 3.67VDD297 pKa = 3.6WYY299 pKa = 11.05RR300 pKa = 11.84DD301 pKa = 3.19PDD303 pKa = 3.73KK304 pKa = 11.26YY305 pKa = 11.27YY306 pKa = 10.84EE307 pKa = 4.43PNLTIGSFQKK317 pKa = 10.71RR318 pKa = 11.84IGTQKK323 pKa = 9.58TVLTALKK330 pKa = 9.92KK331 pKa = 10.5RR332 pKa = 11.84NADD335 pKa = 3.46VPEE338 pKa = 4.18MSDD341 pKa = 3.53AIDD344 pKa = 3.48VKK346 pKa = 10.68RR347 pKa = 11.84VARR350 pKa = 11.84EE351 pKa = 3.74VADD354 pKa = 3.98KK355 pKa = 10.91FISSYY360 pKa = 11.46LNVDD364 pKa = 3.6GKK366 pKa = 10.92DD367 pKa = 3.51CFINGLNVMAKK378 pKa = 10.47GLDD381 pKa = 3.9YY382 pKa = 10.6YY383 pKa = 11.2KK384 pKa = 10.33KK385 pKa = 9.38WKK387 pKa = 9.68DD388 pKa = 3.19HH389 pKa = 6.82RR390 pKa = 11.84EE391 pKa = 3.95LSGVTIATGEE401 pKa = 3.85NLQRR405 pKa = 11.84YY406 pKa = 6.22QHH408 pKa = 6.22MIKK411 pKa = 9.56TDD413 pKa = 3.31IKK415 pKa = 10.77PVVTDD420 pKa = 3.53TLHH423 pKa = 7.11LDD425 pKa = 3.35RR426 pKa = 11.84AIAATITFHH435 pKa = 6.66GKK437 pKa = 10.12GVTSCFSPFFTACFEE452 pKa = 4.22NFSMALKK459 pKa = 10.51SRR461 pKa = 11.84FVVPIGKK468 pKa = 9.37ISSLEE473 pKa = 3.86IPNVSLNNKK482 pKa = 8.32WFLEE486 pKa = 4.07ADD488 pKa = 3.42LSKK491 pKa = 10.63FDD493 pKa = 3.7KK494 pKa = 11.18SQGEE498 pKa = 3.98LHH500 pKa = 7.06LEE502 pKa = 4.06FQRR505 pKa = 11.84EE506 pKa = 3.85ILHH509 pKa = 6.78AIGFPVHH516 pKa = 6.79LSNWWADD523 pKa = 4.13FHH525 pKa = 7.55RR526 pKa = 11.84EE527 pKa = 4.21SYY529 pKa = 11.39LNDD532 pKa = 3.54PNAKK536 pKa = 9.64VSLPVSFQRR545 pKa = 11.84RR546 pKa = 11.84TGDD549 pKa = 2.78AFTYY553 pKa = 10.13FGNTIVTMAMMAYY566 pKa = 8.3TFDD569 pKa = 3.62MNLPEE574 pKa = 4.27LAIFSGDD581 pKa = 3.91DD582 pKa = 3.31SLLVLNDD589 pKa = 3.85KK590 pKa = 10.79PSIDD594 pKa = 3.27TDD596 pKa = 3.19IFQRR600 pKa = 11.84LFNMEE605 pKa = 4.22VKK607 pKa = 10.26IMEE610 pKa = 4.38PSVPYY615 pKa = 10.34VCSKK619 pKa = 10.75FLLEE623 pKa = 4.32TEE625 pKa = 4.41LGSVVSVPDD634 pKa = 4.06PLRR637 pKa = 11.84EE638 pKa = 3.71IQRR641 pKa = 11.84MSKK644 pKa = 10.36RR645 pKa = 11.84KK646 pKa = 8.98ILKK649 pKa = 10.14DD650 pKa = 3.49PEE652 pKa = 3.78VLRR655 pKa = 11.84AHH657 pKa = 6.16FTSFKK662 pKa = 10.89DD663 pKa = 3.25RR664 pKa = 11.84MRR666 pKa = 11.84FLDD669 pKa = 3.56VLDD672 pKa = 4.82DD673 pKa = 4.07RR674 pKa = 11.84MISILCRR681 pKa = 11.84YY682 pKa = 9.56VSLKK686 pKa = 9.64YY687 pKa = 10.43LKK689 pKa = 10.18PGLEE693 pKa = 3.72QDD695 pKa = 3.3VRR697 pKa = 11.84EE698 pKa = 4.21ALACFSYY705 pKa = 10.75YY706 pKa = 10.25SEE708 pKa = 4.31SFFRR712 pKa = 11.84FAEE715 pKa = 5.05LYY717 pKa = 9.82AQDD720 pKa = 3.87GKK722 pKa = 10.95FVYY725 pKa = 9.54QVKK728 pKa = 10.62DD729 pKa = 3.92PISHH733 pKa = 6.4HH734 pKa = 6.7RR735 pKa = 11.84GDD737 pKa = 4.53EE738 pKa = 3.99ILGSRR743 pKa = 11.84EE744 pKa = 3.38KK745 pKa = 11.16DD746 pKa = 2.6GDD748 pKa = 4.06YY749 pKa = 10.72FHH751 pKa = 7.06NWHH754 pKa = 6.49NPVFPRR760 pKa = 11.84ILDD763 pKa = 3.37KK764 pKa = 10.96AVRR767 pKa = 11.84VFGKK771 pKa = 10.68YY772 pKa = 8.11STDD775 pKa = 3.4YY776 pKa = 11.26SSEE779 pKa = 4.06NYY781 pKa = 9.16KK782 pKa = 10.31KK783 pKa = 10.79KK784 pKa = 10.84SMKK787 pKa = 10.5CEE789 pKa = 3.6EE790 pKa = 4.52DD791 pKa = 3.22KK792 pKa = 11.31LFRR795 pKa = 11.84KK796 pKa = 9.73SLSLAYY802 pKa = 9.87DD803 pKa = 3.24QRR805 pKa = 11.84DD806 pKa = 3.43LLKK809 pKa = 11.06NKK811 pKa = 9.74LKK813 pKa = 10.36HH814 pKa = 7.08DD815 pKa = 4.08RR816 pKa = 11.84LLDD819 pKa = 3.62EE820 pKa = 4.64TASSSFSKK828 pKa = 10.95
Molecular weight: 94.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C8KH78|C8KH78_9BROM Coat protein OS=Melandrium yellow fleck virus OX=545259 GN=CP PE=4 SV=1
MM1 pKa = 7.12STPGTVKK8 pKa = 8.18MTRR11 pKa = 11.84AQRR14 pKa = 11.84RR15 pKa = 11.84AAARR19 pKa = 11.84RR20 pKa = 11.84AAKK23 pKa = 9.28PQINKK28 pKa = 9.85RR29 pKa = 11.84IQPVIVEE36 pKa = 4.61PPTSGLGRR44 pKa = 11.84AIKK47 pKa = 10.49ALGGHH52 pKa = 7.13TITKK56 pKa = 9.53WEE58 pKa = 4.33TSCPEE63 pKa = 3.64IKK65 pKa = 10.31AKK67 pKa = 10.64SSIQITIPLPSEE79 pKa = 4.05LSSEE83 pKa = 4.13RR84 pKa = 11.84NKK86 pKa = 9.91VLKK89 pKa = 10.31VGRR92 pKa = 11.84VLLWLGLLPSINGQVKK108 pKa = 10.43ACVTQTNPVPGAAFQSALAIADD130 pKa = 3.47SSKK133 pKa = 10.73EE134 pKa = 4.18VVSAMYY140 pKa = 10.85VDD142 pKa = 3.94IFKK145 pKa = 11.19GITIEE150 pKa = 5.52DD151 pKa = 4.12FVNDD155 pKa = 3.7LAIYY159 pKa = 9.59VYY161 pKa = 10.74SAVDD165 pKa = 3.36VAAKK169 pKa = 10.3DD170 pKa = 3.64IIVHH174 pKa = 6.09LEE176 pKa = 3.96VEE178 pKa = 4.46HH179 pKa = 5.9VRR181 pKa = 11.84PVLDD185 pKa = 5.11DD186 pKa = 3.74YY187 pKa = 9.14FTPLHH192 pKa = 6.2
MM1 pKa = 7.12STPGTVKK8 pKa = 8.18MTRR11 pKa = 11.84AQRR14 pKa = 11.84RR15 pKa = 11.84AAARR19 pKa = 11.84RR20 pKa = 11.84AAKK23 pKa = 9.28PQINKK28 pKa = 9.85RR29 pKa = 11.84IQPVIVEE36 pKa = 4.61PPTSGLGRR44 pKa = 11.84AIKK47 pKa = 10.49ALGGHH52 pKa = 7.13TITKK56 pKa = 9.53WEE58 pKa = 4.33TSCPEE63 pKa = 3.64IKK65 pKa = 10.31AKK67 pKa = 10.64SSIQITIPLPSEE79 pKa = 4.05LSSEE83 pKa = 4.13RR84 pKa = 11.84NKK86 pKa = 9.91VLKK89 pKa = 10.31VGRR92 pKa = 11.84VLLWLGLLPSINGQVKK108 pKa = 10.43ACVTQTNPVPGAAFQSALAIADD130 pKa = 3.47SSKK133 pKa = 10.73EE134 pKa = 4.18VVSAMYY140 pKa = 10.85VDD142 pKa = 3.94IFKK145 pKa = 11.19GITIEE150 pKa = 5.52DD151 pKa = 4.12FVNDD155 pKa = 3.7LAIYY159 pKa = 9.59VYY161 pKa = 10.74SAVDD165 pKa = 3.36VAAKK169 pKa = 10.3DD170 pKa = 3.64IIVHH174 pKa = 6.09LEE176 pKa = 3.96VEE178 pKa = 4.46HH179 pKa = 5.9VRR181 pKa = 11.84PVLDD185 pKa = 5.11DD186 pKa = 3.74YY187 pKa = 9.14FTPLHH192 pKa = 6.2
Molecular weight: 20.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2263 |
192 |
948 |
565.8 |
63.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.275 ± 0.702 | 2.121 ± 0.694 |
7.114 ± 1.188 | 6.142 ± 0.552 |
4.242 ± 0.692 | 5.259 ± 0.425 |
2.342 ± 0.352 | 5.259 ± 0.691 |
6.894 ± 0.125 | 8.484 ± 0.545 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.298 ± 0.121 | 4.596 ± 0.507 |
4.11 ± 0.885 | 2.872 ± 0.165 |
5.435 ± 0.566 | 8.175 ± 1.148 |
5.038 ± 0.4 | 8.263 ± 0.685 |
1.326 ± 0.221 | 3.756 ± 0.412 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
