
Streptomyces sp. 8K308
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
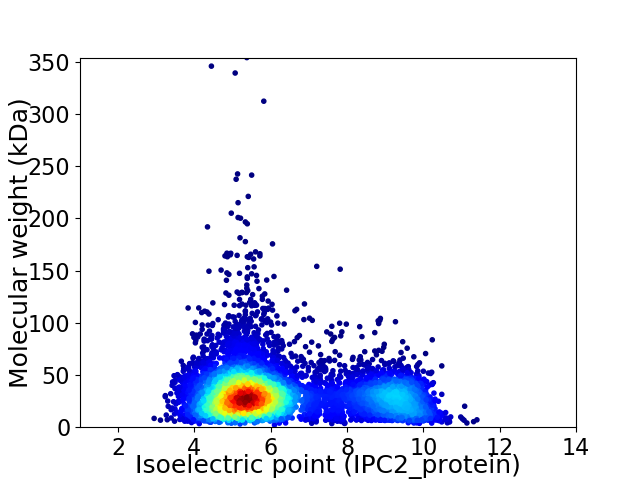
Virtual 2D-PAGE plot for 6672 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
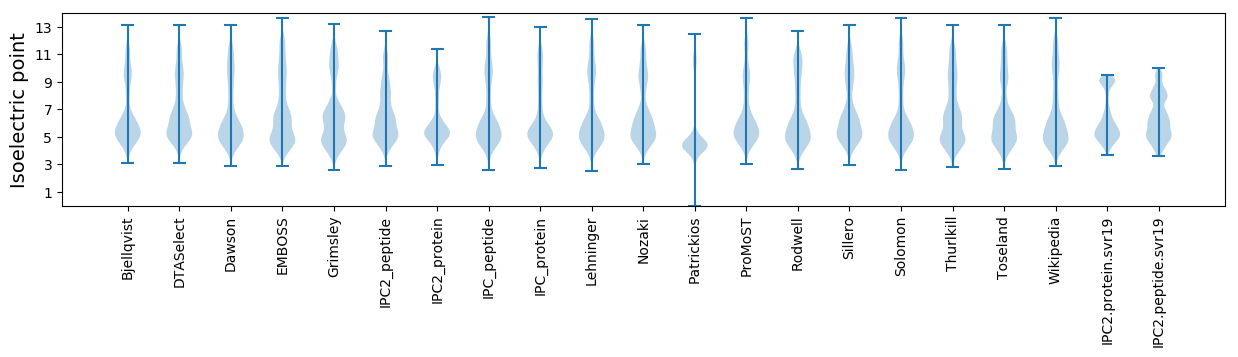
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R4PL97|A0A4R4PL97_9ACTN Glycoside hydrolase OS=Streptomyces sp. 8K308 OX=2530388 GN=E1265_14805 PE=3 SV=1
MM1 pKa = 7.73RR2 pKa = 11.84GARR5 pKa = 11.84SAKK8 pKa = 8.19WVVSAIVVAMAATACGSDD26 pKa = 3.77SDD28 pKa = 5.22DD29 pKa = 4.37NGDD32 pKa = 3.69GGASGEE38 pKa = 4.3GNSAGIVRR46 pKa = 11.84VDD48 pKa = 3.27GGEE51 pKa = 4.01PQNPLIPTNTTEE63 pKa = 3.86QYY65 pKa = 10.31GALVIQNVWSKK76 pKa = 11.06LVDD79 pKa = 4.0FDD81 pKa = 5.48DD82 pKa = 3.76QGQVFTVAAEE92 pKa = 4.18SVEE95 pKa = 4.22PNEE98 pKa = 4.83DD99 pKa = 3.17ASVWTVTLKK108 pKa = 10.99DD109 pKa = 2.91GWSFHH114 pKa = 6.3NGEE117 pKa = 4.3AVTAQSYY124 pKa = 9.45VDD126 pKa = 3.38AWNWAANVNNNQQNSFWFSDD146 pKa = 2.9IAGYY150 pKa = 10.92EE151 pKa = 4.04DD152 pKa = 3.51VHH154 pKa = 8.1PEE156 pKa = 3.91EE157 pKa = 5.41GEE159 pKa = 4.08PTADD163 pKa = 3.03TMSGLEE169 pKa = 4.11VVDD172 pKa = 5.45DD173 pKa = 3.96LTFTITLNTPQSYY186 pKa = 8.71YY187 pKa = 10.91DD188 pKa = 3.47YY189 pKa = 10.98RR190 pKa = 11.84LGYY193 pKa = 10.28DD194 pKa = 3.83AFAPLPSVFYY204 pKa = 10.39DD205 pKa = 3.51DD206 pKa = 4.88PEE208 pKa = 5.64AFGEE212 pKa = 4.26HH213 pKa = 7.31PIGNGPYY220 pKa = 10.52EE221 pKa = 4.05FTEE224 pKa = 4.23WNHH227 pKa = 5.74NEE229 pKa = 4.39SISLTAWDD237 pKa = 5.32EE238 pKa = 4.24YY239 pKa = 10.97PGDD242 pKa = 4.52DD243 pKa = 3.75KK244 pKa = 11.22PANGGVEE251 pKa = 3.92LVAYY255 pKa = 9.86DD256 pKa = 4.27NLHH259 pKa = 5.96TAYY262 pKa = 10.48QDD264 pKa = 3.88LLSGNLDD271 pKa = 3.36VLRR274 pKa = 11.84EE275 pKa = 3.85VDD277 pKa = 3.84AQDD280 pKa = 3.62LPVYY284 pKa = 8.81EE285 pKa = 4.45TDD287 pKa = 3.66LGDD290 pKa = 3.51RR291 pKa = 11.84AIAQPYY297 pKa = 10.19NGTQSIVPAFYY308 pKa = 10.34SDD310 pKa = 3.41EE311 pKa = 4.6FEE313 pKa = 5.3DD314 pKa = 3.31VDD316 pKa = 3.88PRR318 pKa = 11.84VLQGMSLAIDD328 pKa = 3.68RR329 pKa = 11.84EE330 pKa = 4.73TITGTVLNGSATPADD345 pKa = 3.99SFVAPGVFGYY355 pKa = 10.15QEE357 pKa = 4.54GIGAEE362 pKa = 4.15FTAYY366 pKa = 10.28DD367 pKa = 3.73PEE369 pKa = 4.23RR370 pKa = 11.84ARR372 pKa = 11.84EE373 pKa = 3.96LFEE376 pKa = 6.12AGGGIPGNTVTIQYY390 pKa = 10.11NADD393 pKa = 3.32GPHH396 pKa = 6.35RR397 pKa = 11.84EE398 pKa = 4.05WVEE401 pKa = 3.9AVCGNLRR408 pKa = 11.84DD409 pKa = 4.03NLGVEE414 pKa = 4.73CEE416 pKa = 4.21TDD418 pKa = 3.63VKK420 pKa = 11.04PDD422 pKa = 3.6FATDD426 pKa = 3.79LEE428 pKa = 4.4ARR430 pKa = 11.84DD431 pKa = 3.62ADD433 pKa = 4.19EE434 pKa = 3.85VVGMYY439 pKa = 10.28RR440 pKa = 11.84GGWFQDD446 pKa = 3.39YY447 pKa = 9.82PLNVNFLRR455 pKa = 11.84EE456 pKa = 4.37LYY458 pKa = 9.02RR459 pKa = 11.84TGASTNYY466 pKa = 9.4GRR468 pKa = 11.84YY469 pKa = 9.79SNEE472 pKa = 3.49EE473 pKa = 3.4VDD475 pKa = 4.48ALFAEE480 pKa = 5.19GDD482 pKa = 3.76SSASLDD488 pKa = 3.75DD489 pKa = 3.52TVAAYY494 pKa = 10.14QEE496 pKa = 4.38AEE498 pKa = 3.99EE499 pKa = 5.61LLFQDD504 pKa = 4.56MPAIPLWFQNINGGYY519 pKa = 9.14SEE521 pKa = 5.21NVDD524 pKa = 3.1NVRR527 pKa = 11.84FDD529 pKa = 3.4MAGQPVLTEE538 pKa = 3.89VTVNN542 pKa = 3.26
MM1 pKa = 7.73RR2 pKa = 11.84GARR5 pKa = 11.84SAKK8 pKa = 8.19WVVSAIVVAMAATACGSDD26 pKa = 3.77SDD28 pKa = 5.22DD29 pKa = 4.37NGDD32 pKa = 3.69GGASGEE38 pKa = 4.3GNSAGIVRR46 pKa = 11.84VDD48 pKa = 3.27GGEE51 pKa = 4.01PQNPLIPTNTTEE63 pKa = 3.86QYY65 pKa = 10.31GALVIQNVWSKK76 pKa = 11.06LVDD79 pKa = 4.0FDD81 pKa = 5.48DD82 pKa = 3.76QGQVFTVAAEE92 pKa = 4.18SVEE95 pKa = 4.22PNEE98 pKa = 4.83DD99 pKa = 3.17ASVWTVTLKK108 pKa = 10.99DD109 pKa = 2.91GWSFHH114 pKa = 6.3NGEE117 pKa = 4.3AVTAQSYY124 pKa = 9.45VDD126 pKa = 3.38AWNWAANVNNNQQNSFWFSDD146 pKa = 2.9IAGYY150 pKa = 10.92EE151 pKa = 4.04DD152 pKa = 3.51VHH154 pKa = 8.1PEE156 pKa = 3.91EE157 pKa = 5.41GEE159 pKa = 4.08PTADD163 pKa = 3.03TMSGLEE169 pKa = 4.11VVDD172 pKa = 5.45DD173 pKa = 3.96LTFTITLNTPQSYY186 pKa = 8.71YY187 pKa = 10.91DD188 pKa = 3.47YY189 pKa = 10.98RR190 pKa = 11.84LGYY193 pKa = 10.28DD194 pKa = 3.83AFAPLPSVFYY204 pKa = 10.39DD205 pKa = 3.51DD206 pKa = 4.88PEE208 pKa = 5.64AFGEE212 pKa = 4.26HH213 pKa = 7.31PIGNGPYY220 pKa = 10.52EE221 pKa = 4.05FTEE224 pKa = 4.23WNHH227 pKa = 5.74NEE229 pKa = 4.39SISLTAWDD237 pKa = 5.32EE238 pKa = 4.24YY239 pKa = 10.97PGDD242 pKa = 4.52DD243 pKa = 3.75KK244 pKa = 11.22PANGGVEE251 pKa = 3.92LVAYY255 pKa = 9.86DD256 pKa = 4.27NLHH259 pKa = 5.96TAYY262 pKa = 10.48QDD264 pKa = 3.88LLSGNLDD271 pKa = 3.36VLRR274 pKa = 11.84EE275 pKa = 3.85VDD277 pKa = 3.84AQDD280 pKa = 3.62LPVYY284 pKa = 8.81EE285 pKa = 4.45TDD287 pKa = 3.66LGDD290 pKa = 3.51RR291 pKa = 11.84AIAQPYY297 pKa = 10.19NGTQSIVPAFYY308 pKa = 10.34SDD310 pKa = 3.41EE311 pKa = 4.6FEE313 pKa = 5.3DD314 pKa = 3.31VDD316 pKa = 3.88PRR318 pKa = 11.84VLQGMSLAIDD328 pKa = 3.68RR329 pKa = 11.84EE330 pKa = 4.73TITGTVLNGSATPADD345 pKa = 3.99SFVAPGVFGYY355 pKa = 10.15QEE357 pKa = 4.54GIGAEE362 pKa = 4.15FTAYY366 pKa = 10.28DD367 pKa = 3.73PEE369 pKa = 4.23RR370 pKa = 11.84ARR372 pKa = 11.84EE373 pKa = 3.96LFEE376 pKa = 6.12AGGGIPGNTVTIQYY390 pKa = 10.11NADD393 pKa = 3.32GPHH396 pKa = 6.35RR397 pKa = 11.84EE398 pKa = 4.05WVEE401 pKa = 3.9AVCGNLRR408 pKa = 11.84DD409 pKa = 4.03NLGVEE414 pKa = 4.73CEE416 pKa = 4.21TDD418 pKa = 3.63VKK420 pKa = 11.04PDD422 pKa = 3.6FATDD426 pKa = 3.79LEE428 pKa = 4.4ARR430 pKa = 11.84DD431 pKa = 3.62ADD433 pKa = 4.19EE434 pKa = 3.85VVGMYY439 pKa = 10.28RR440 pKa = 11.84GGWFQDD446 pKa = 3.39YY447 pKa = 9.82PLNVNFLRR455 pKa = 11.84EE456 pKa = 4.37LYY458 pKa = 9.02RR459 pKa = 11.84TGASTNYY466 pKa = 9.4GRR468 pKa = 11.84YY469 pKa = 9.79SNEE472 pKa = 3.49EE473 pKa = 3.4VDD475 pKa = 4.48ALFAEE480 pKa = 5.19GDD482 pKa = 3.76SSASLDD488 pKa = 3.75DD489 pKa = 3.52TVAAYY494 pKa = 10.14QEE496 pKa = 4.38AEE498 pKa = 3.99EE499 pKa = 5.61LLFQDD504 pKa = 4.56MPAIPLWFQNINGGYY519 pKa = 9.14SEE521 pKa = 5.21NVDD524 pKa = 3.1NVRR527 pKa = 11.84FDD529 pKa = 3.4MAGQPVLTEE538 pKa = 3.89VTVNN542 pKa = 3.26
Molecular weight: 59.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R4NTH5|A0A4R4NTH5_9ACTN PAS domain-containing protein (Fragment) OS=Streptomyces sp. 8K308 OX=2530388 GN=E1265_30100 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIASRR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.43GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIASRR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.43GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2115711 |
18 |
3386 |
317.1 |
33.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.063 ± 0.054 | 0.758 ± 0.009 |
6.001 ± 0.026 | 6.025 ± 0.031 |
2.679 ± 0.017 | 9.483 ± 0.025 |
2.288 ± 0.013 | 3.069 ± 0.022 |
1.293 ± 0.02 | 10.674 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.631 ± 0.013 | 1.719 ± 0.017 |
6.208 ± 0.024 | 2.576 ± 0.017 |
8.8 ± 0.035 | 4.75 ± 0.023 |
6.008 ± 0.027 | 8.402 ± 0.027 |
1.578 ± 0.013 | 1.997 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
