
Sphingomonas sp. HDW15C
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
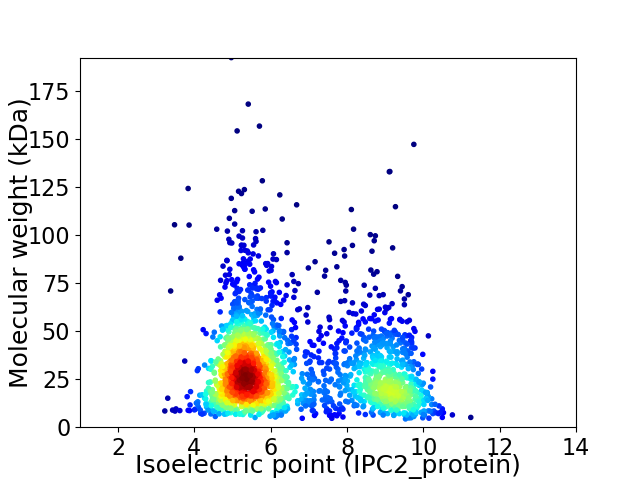
Virtual 2D-PAGE plot for 2087 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
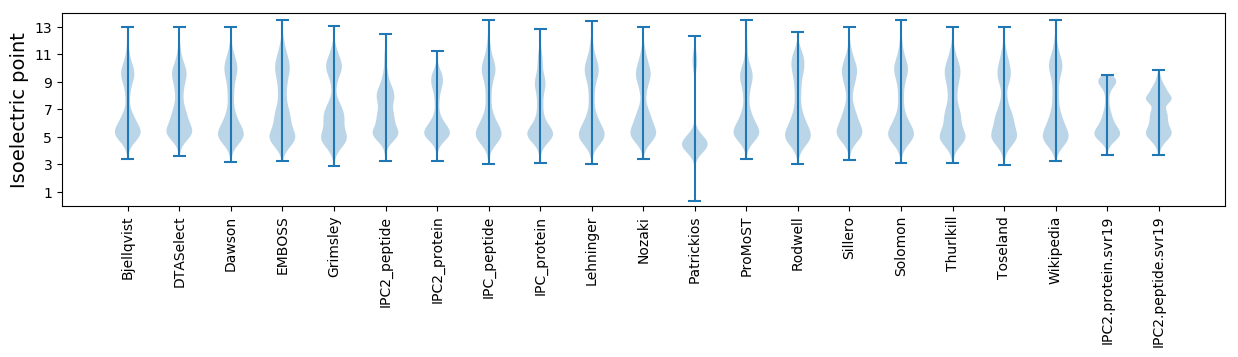
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G7ZLN4|A0A6G7ZLN4_9SPHN RNA polymerase sigma factor OS=Sphingomonas sp. HDW15C OX=2714944 GN=G7078_02855 PE=3 SV=1
MM1 pKa = 7.81VDD3 pKa = 3.21IPGDD7 pKa = 3.6TTTTSTISVGSMVDD21 pKa = 3.38NAIEE25 pKa = 4.19TVGDD29 pKa = 3.95HH30 pKa = 6.99DD31 pKa = 4.31WYY33 pKa = 11.04AVTLTAGQKK42 pKa = 8.01VTIALNIITLEE53 pKa = 4.09DD54 pKa = 4.07PYY56 pKa = 11.56LYY58 pKa = 10.87LRR60 pKa = 11.84DD61 pKa = 4.08SNGSLIAEE69 pKa = 4.4NDD71 pKa = 3.47DD72 pKa = 3.84GGGGRR77 pKa = 11.84GSRR80 pKa = 11.84LVFTAPTDD88 pKa = 3.31GTYY91 pKa = 10.85YY92 pKa = 10.24IDD94 pKa = 3.57VAAWAPTEE102 pKa = 4.06VVPGYY107 pKa = 8.99TGTGAYY113 pKa = 9.34RR114 pKa = 11.84LSVSDD119 pKa = 3.72YY120 pKa = 9.02VAPSEE125 pKa = 4.2GTLDD129 pKa = 4.42DD130 pKa = 5.06FADD133 pKa = 3.66QMTHH137 pKa = 6.01GFFDD141 pKa = 3.86GDD143 pKa = 3.59YY144 pKa = 10.03HH145 pKa = 7.28HH146 pKa = 7.35FNVTQGGSLTVNFQGLTDD164 pKa = 4.17AGRR167 pKa = 11.84TVALQALQQWTDD179 pKa = 3.15IIGVNFVEE187 pKa = 4.72TNGAAQITFDD197 pKa = 5.75DD198 pKa = 4.67LDD200 pKa = 3.63QGTGAFADD208 pKa = 4.13TVSSNHH214 pKa = 5.36ITSSAIVNVALYY226 pKa = 10.29RR227 pKa = 11.84LNLHH231 pKa = 6.45TYY233 pKa = 6.25MHH235 pKa = 7.43EE236 pKa = 3.98IGHH239 pKa = 6.67ALGIGHH245 pKa = 7.1TSNSNAGTAGAIYY258 pKa = 10.09PNDD261 pKa = 4.07ALWSNDD267 pKa = 2.34GSAISIMSYY276 pKa = 10.72FDD278 pKa = 3.3NGEE281 pKa = 3.75NGYY284 pKa = 10.0YY285 pKa = 10.0SSRR288 pKa = 11.84GFSNLPVVTPQVADD302 pKa = 4.57IIAMGNLYY310 pKa = 10.61GLSTTTRR317 pKa = 11.84TGNTTYY323 pKa = 11.17GFNNTSGRR331 pKa = 11.84AAFDD335 pKa = 3.54AALHH339 pKa = 6.09PNYY342 pKa = 10.49SYY344 pKa = 11.07TIFDD348 pKa = 3.57NGGVDD353 pKa = 3.31TLDD356 pKa = 3.28YY357 pKa = 11.41SGFISNQLIDD367 pKa = 4.73LNPEE371 pKa = 3.71AFSNIGAFVGNVVIARR387 pKa = 11.84GTVIEE392 pKa = 4.05NAIGGSGNDD401 pKa = 3.41TLVGNGAANVLTGNSGIDD419 pKa = 3.34TLYY422 pKa = 11.17GADD425 pKa = 4.46GNDD428 pKa = 3.49TLTGGLGADD437 pKa = 3.56VLVGGTGGDD446 pKa = 3.87FFRR449 pKa = 11.84DD450 pKa = 3.28TAAGLNGDD458 pKa = 4.28TIVDD462 pKa = 4.45FSAQDD467 pKa = 3.42RR468 pKa = 11.84IVITNVGLASFSYY481 pKa = 10.68SLVGNTLTYY490 pKa = 10.26TGGSLTFSNALAGHH504 pKa = 6.67LFVRR508 pKa = 11.84AADD511 pKa = 3.79GGGVEE516 pKa = 4.92LQMADD521 pKa = 3.0RR522 pKa = 11.84TGFGLTDD529 pKa = 3.79FNGDD533 pKa = 4.13GIADD537 pKa = 3.88VLWRR541 pKa = 11.84SDD543 pKa = 3.23TGVITTWLGQADD555 pKa = 4.13GTFIDD560 pKa = 4.07NSANTGQAIPLDD572 pKa = 3.92WNIVGTGDD580 pKa = 3.67YY581 pKa = 11.04NGDD584 pKa = 3.62GLGDD588 pKa = 3.57IMWRR592 pKa = 11.84SDD594 pKa = 3.85SGVMTQWLGQLDD606 pKa = 3.93GTFRR610 pKa = 11.84DD611 pKa = 3.9NFAKK615 pKa = 9.74TGQVIPLDD623 pKa = 3.61WNVIGTGDD631 pKa = 3.78FNGDD635 pKa = 3.42GLGDD639 pKa = 4.64LIWRR643 pKa = 11.84SDD645 pKa = 3.26AGVITEE651 pKa = 4.32WLGNLDD657 pKa = 4.03GSFRR661 pKa = 11.84DD662 pKa = 3.79NFAGTGQHH670 pKa = 6.8IPLNWTIDD678 pKa = 3.28GMGDD682 pKa = 3.48FNGDD686 pKa = 3.51GLEE689 pKa = 3.97DD690 pKa = 4.84LIWRR694 pKa = 11.84SDD696 pKa = 3.19AGVITEE702 pKa = 4.5WLGQNDD708 pKa = 4.51GSFKK712 pKa = 11.28DD713 pKa = 3.74NFAGTGQTIPTNWQIVGVGDD733 pKa = 4.48FNADD737 pKa = 3.17GYY739 pKa = 11.89ADD741 pKa = 4.33ILWQSTAGTLTNWLGNPDD759 pKa = 4.32GSFTDD764 pKa = 3.56NFVNTGQVIPQGWEE778 pKa = 3.63LVGVGDD784 pKa = 4.91LNGDD788 pKa = 3.51GRR790 pKa = 11.84DD791 pKa = 3.85DD792 pKa = 4.25ILWQTGPDD800 pKa = 3.65SVEE803 pKa = 3.58TWFGQDD809 pKa = 2.52NGGFTGGSSGTLAAAHH825 pKa = 6.74AATAGTNGDD834 pKa = 4.03LFYY837 pKa = 11.18LGG839 pKa = 4.25
MM1 pKa = 7.81VDD3 pKa = 3.21IPGDD7 pKa = 3.6TTTTSTISVGSMVDD21 pKa = 3.38NAIEE25 pKa = 4.19TVGDD29 pKa = 3.95HH30 pKa = 6.99DD31 pKa = 4.31WYY33 pKa = 11.04AVTLTAGQKK42 pKa = 8.01VTIALNIITLEE53 pKa = 4.09DD54 pKa = 4.07PYY56 pKa = 11.56LYY58 pKa = 10.87LRR60 pKa = 11.84DD61 pKa = 4.08SNGSLIAEE69 pKa = 4.4NDD71 pKa = 3.47DD72 pKa = 3.84GGGGRR77 pKa = 11.84GSRR80 pKa = 11.84LVFTAPTDD88 pKa = 3.31GTYY91 pKa = 10.85YY92 pKa = 10.24IDD94 pKa = 3.57VAAWAPTEE102 pKa = 4.06VVPGYY107 pKa = 8.99TGTGAYY113 pKa = 9.34RR114 pKa = 11.84LSVSDD119 pKa = 3.72YY120 pKa = 9.02VAPSEE125 pKa = 4.2GTLDD129 pKa = 4.42DD130 pKa = 5.06FADD133 pKa = 3.66QMTHH137 pKa = 6.01GFFDD141 pKa = 3.86GDD143 pKa = 3.59YY144 pKa = 10.03HH145 pKa = 7.28HH146 pKa = 7.35FNVTQGGSLTVNFQGLTDD164 pKa = 4.17AGRR167 pKa = 11.84TVALQALQQWTDD179 pKa = 3.15IIGVNFVEE187 pKa = 4.72TNGAAQITFDD197 pKa = 5.75DD198 pKa = 4.67LDD200 pKa = 3.63QGTGAFADD208 pKa = 4.13TVSSNHH214 pKa = 5.36ITSSAIVNVALYY226 pKa = 10.29RR227 pKa = 11.84LNLHH231 pKa = 6.45TYY233 pKa = 6.25MHH235 pKa = 7.43EE236 pKa = 3.98IGHH239 pKa = 6.67ALGIGHH245 pKa = 7.1TSNSNAGTAGAIYY258 pKa = 10.09PNDD261 pKa = 4.07ALWSNDD267 pKa = 2.34GSAISIMSYY276 pKa = 10.72FDD278 pKa = 3.3NGEE281 pKa = 3.75NGYY284 pKa = 10.0YY285 pKa = 10.0SSRR288 pKa = 11.84GFSNLPVVTPQVADD302 pKa = 4.57IIAMGNLYY310 pKa = 10.61GLSTTTRR317 pKa = 11.84TGNTTYY323 pKa = 11.17GFNNTSGRR331 pKa = 11.84AAFDD335 pKa = 3.54AALHH339 pKa = 6.09PNYY342 pKa = 10.49SYY344 pKa = 11.07TIFDD348 pKa = 3.57NGGVDD353 pKa = 3.31TLDD356 pKa = 3.28YY357 pKa = 11.41SGFISNQLIDD367 pKa = 4.73LNPEE371 pKa = 3.71AFSNIGAFVGNVVIARR387 pKa = 11.84GTVIEE392 pKa = 4.05NAIGGSGNDD401 pKa = 3.41TLVGNGAANVLTGNSGIDD419 pKa = 3.34TLYY422 pKa = 11.17GADD425 pKa = 4.46GNDD428 pKa = 3.49TLTGGLGADD437 pKa = 3.56VLVGGTGGDD446 pKa = 3.87FFRR449 pKa = 11.84DD450 pKa = 3.28TAAGLNGDD458 pKa = 4.28TIVDD462 pKa = 4.45FSAQDD467 pKa = 3.42RR468 pKa = 11.84IVITNVGLASFSYY481 pKa = 10.68SLVGNTLTYY490 pKa = 10.26TGGSLTFSNALAGHH504 pKa = 6.67LFVRR508 pKa = 11.84AADD511 pKa = 3.79GGGVEE516 pKa = 4.92LQMADD521 pKa = 3.0RR522 pKa = 11.84TGFGLTDD529 pKa = 3.79FNGDD533 pKa = 4.13GIADD537 pKa = 3.88VLWRR541 pKa = 11.84SDD543 pKa = 3.23TGVITTWLGQADD555 pKa = 4.13GTFIDD560 pKa = 4.07NSANTGQAIPLDD572 pKa = 3.92WNIVGTGDD580 pKa = 3.67YY581 pKa = 11.04NGDD584 pKa = 3.62GLGDD588 pKa = 3.57IMWRR592 pKa = 11.84SDD594 pKa = 3.85SGVMTQWLGQLDD606 pKa = 3.93GTFRR610 pKa = 11.84DD611 pKa = 3.9NFAKK615 pKa = 9.74TGQVIPLDD623 pKa = 3.61WNVIGTGDD631 pKa = 3.78FNGDD635 pKa = 3.42GLGDD639 pKa = 4.64LIWRR643 pKa = 11.84SDD645 pKa = 3.26AGVITEE651 pKa = 4.32WLGNLDD657 pKa = 4.03GSFRR661 pKa = 11.84DD662 pKa = 3.79NFAGTGQHH670 pKa = 6.8IPLNWTIDD678 pKa = 3.28GMGDD682 pKa = 3.48FNGDD686 pKa = 3.51GLEE689 pKa = 3.97DD690 pKa = 4.84LIWRR694 pKa = 11.84SDD696 pKa = 3.19AGVITEE702 pKa = 4.5WLGQNDD708 pKa = 4.51GSFKK712 pKa = 11.28DD713 pKa = 3.74NFAGTGQTIPTNWQIVGVGDD733 pKa = 4.48FNADD737 pKa = 3.17GYY739 pKa = 11.89ADD741 pKa = 4.33ILWQSTAGTLTNWLGNPDD759 pKa = 4.32GSFTDD764 pKa = 3.56NFVNTGQVIPQGWEE778 pKa = 3.63LVGVGDD784 pKa = 4.91LNGDD788 pKa = 3.51GRR790 pKa = 11.84DD791 pKa = 3.85DD792 pKa = 4.25ILWQTGPDD800 pKa = 3.65SVEE803 pKa = 3.58TWFGQDD809 pKa = 2.52NGGFTGGSSGTLAAAHH825 pKa = 6.74AATAGTNGDD834 pKa = 4.03LFYY837 pKa = 11.18LGG839 pKa = 4.25
Molecular weight: 88.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G7ZQW8|A0A6G7ZQW8_9SPHN Probable multidrug resistance protein NorM OS=Sphingomonas sp. HDW15C OX=2714944 GN=G7078_04370 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.08VLNARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84NKK41 pKa = 10.44LSAA44 pKa = 3.94
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.08VLNARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84NKK41 pKa = 10.44LSAA44 pKa = 3.94
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
640540 |
41 |
1791 |
306.9 |
33.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.1 ± 0.083 | 0.761 ± 0.016 |
5.913 ± 0.045 | 5.748 ± 0.057 |
3.541 ± 0.035 | 8.84 ± 0.052 |
1.836 ± 0.024 | 4.923 ± 0.036 |
3.261 ± 0.044 | 10.0 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.272 ± 0.023 | 2.495 ± 0.029 |
5.334 ± 0.042 | 3.285 ± 0.03 |
7.537 ± 0.057 | 5.391 ± 0.036 |
4.944 ± 0.038 | 7.352 ± 0.044 |
1.372 ± 0.025 | 2.096 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
