
Candidatus Marinimicrobia bacterium PRS2
Taxonomy: cellular organisms; Bacteria; FCB group; Candidatus Marinimicrobia
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
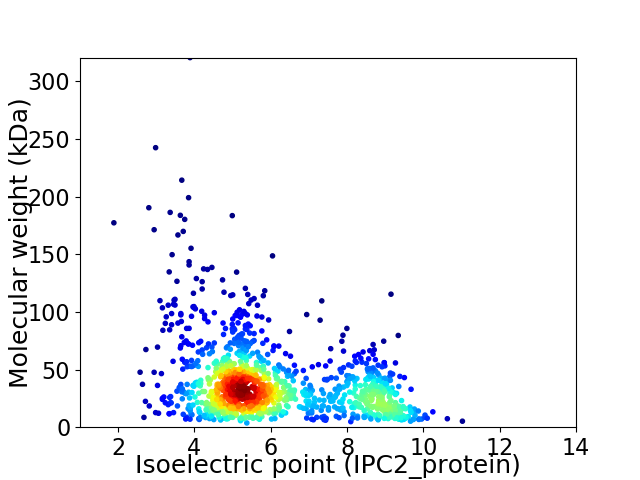
Virtual 2D-PAGE plot for 1209 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3M7KIF0|A0A3M7KIF0_9BACT Uncharacterized protein OS=Candidatus Marinimicrobia bacterium PRS2 OX=2483458 GN=EB821_05875 PE=4 SV=1
SS1 pKa = 6.79NGGLGGSTVIATVTVNPVNDD21 pKa = 3.86EE22 pKa = 4.03PSFEE26 pKa = 4.28YY27 pKa = 10.92TNVVAVNEE35 pKa = 4.05DD36 pKa = 3.32SGLYY40 pKa = 7.63EE41 pKa = 4.5ASWAFNISVGPDD53 pKa = 3.25NEE55 pKa = 4.47SDD57 pKa = 3.59QIPTFEE63 pKa = 5.38IISNDD68 pKa = 3.2HH69 pKa = 6.33EE70 pKa = 5.51SYY72 pKa = 10.98FSIQPSISEE81 pKa = 4.23LGVLTFTPADD91 pKa = 3.8NQHH94 pKa = 6.04GEE96 pKa = 4.09AIITVRR102 pKa = 11.84LTDD105 pKa = 3.55NGGTEE110 pKa = 3.97NGGVNTYY117 pKa = 11.09DD118 pKa = 3.9DD119 pKa = 3.41QTFTIKK125 pKa = 10.63LNPIADD131 pKa = 4.08TPNITDD137 pKa = 3.81AEE139 pKa = 4.16TDD141 pKa = 3.42EE142 pKa = 6.81DD143 pKa = 4.04IMSSSGLVITRR154 pKa = 11.84NAVDD158 pKa = 3.77SEE160 pKa = 4.52EE161 pKa = 4.12VTHH164 pKa = 7.03FKK166 pKa = 9.2ITGITNGTLYY176 pKa = 10.68QNNGSTSIANNEE188 pKa = 4.26FITFTEE194 pKa = 4.74GNTGLKK200 pKa = 9.9FDD202 pKa = 5.24PDD204 pKa = 3.11EE205 pKa = 5.15DD206 pKa = 5.24YY207 pKa = 11.3YY208 pKa = 11.73GSGFITIQASVSSSNGGLGGSTVIATVTVNPEE240 pKa = 3.41NDD242 pKa = 3.5GPYY245 pKa = 10.26DD246 pKa = 3.8AEE248 pKa = 6.0AGDD251 pKa = 5.2DD252 pKa = 4.07IYY254 pKa = 11.41DD255 pKa = 4.15ILPSDD260 pKa = 3.82ATTIDD265 pKa = 3.28ITILDD270 pKa = 4.11VSSDD274 pKa = 4.41DD275 pKa = 3.66IDD277 pKa = 6.61DD278 pKa = 4.54DD279 pKa = 4.82VSVSHH284 pKa = 6.76TYY286 pKa = 10.1SWTLIDD292 pKa = 5.1ANNGDD297 pKa = 4.61IIAEE301 pKa = 3.98ATEE304 pKa = 4.06INPTFALKK312 pKa = 10.39AGHH315 pKa = 5.79YY316 pKa = 8.9QFEE319 pKa = 4.7LTVTDD324 pKa = 3.49TTTNDD329 pKa = 2.94SNIYY333 pKa = 10.86GDD335 pKa = 3.52ITDD338 pKa = 4.11TDD340 pKa = 4.02TIDD343 pKa = 3.76IYY345 pKa = 10.68IGQPGVDD352 pKa = 3.08ISSNPVFVTYY362 pKa = 11.05DD363 pKa = 3.28DD364 pKa = 3.34VAYY367 pKa = 9.94IDD369 pKa = 3.32IVYY372 pKa = 10.27FEE374 pKa = 5.05HH375 pKa = 7.44EE376 pKa = 4.54YY377 pKa = 11.46ASVTQNGDD385 pKa = 3.37QIVISVPIEE394 pKa = 3.97YY395 pKa = 10.11QNKK398 pKa = 7.66FQFIAGQIIPQDD410 pKa = 3.42ALEE413 pKa = 4.08IQIGYY418 pKa = 9.65SGDD421 pKa = 3.42VTDD424 pKa = 5.71ISIEE428 pKa = 4.32SITTSAITINVVTSHH443 pKa = 6.41ISEE446 pKa = 3.99NFYY449 pKa = 10.82IKK451 pKa = 10.57LKK453 pKa = 10.19SVEE456 pKa = 4.19VNPIDD461 pKa = 3.74VTSDD465 pKa = 3.41FSLEE469 pKa = 4.06IEE471 pKa = 4.22NDD473 pKa = 3.03NYY475 pKa = 11.46LPILKK480 pKa = 9.91SATEE484 pKa = 3.82ISIGNPKK491 pKa = 8.31ITTITDD497 pKa = 3.35QVLITANPNEE507 pKa = 4.23KK508 pKa = 9.44MLDD511 pKa = 3.35IIVYY515 pKa = 9.56EE516 pKa = 4.46DD517 pKa = 3.7EE518 pKa = 4.34SVAAINTDD526 pKa = 3.61DD527 pKa = 5.84DD528 pKa = 3.95IVIQFPSTLEE538 pKa = 4.15NNVKK542 pKa = 9.8WIQVPTASSNSEE554 pKa = 4.1KK555 pKa = 10.49IGTITIDD562 pKa = 3.38SSDD565 pKa = 3.77STKK568 pKa = 11.16LLIDD572 pKa = 4.36VISDD576 pKa = 3.35FFAGEE581 pKa = 4.18SLILTDD587 pKa = 5.28GYY589 pKa = 11.02VQVGDD594 pKa = 3.89IEE596 pKa = 5.24SNEE599 pKa = 3.71DD600 pKa = 3.26DD601 pKa = 4.07YY602 pKa = 12.12YY603 pKa = 11.59KK604 pKa = 10.54IIAYY608 pKa = 10.21VNGRR612 pKa = 11.84NQSLDD617 pKa = 3.61VEE619 pKa = 4.76CDD621 pKa = 2.8NSIIVTSPEE630 pKa = 3.59LSFASDD636 pKa = 2.57QHH638 pKa = 6.88YY639 pKa = 10.64VVNDD643 pKa = 3.61SAVSFPTITLTEE655 pKa = 4.1SSLIPSIYY663 pKa = 10.53SEE665 pKa = 4.35VQDD668 pKa = 4.23SIYY671 pKa = 10.84LRR673 pKa = 11.84FDD675 pKa = 3.68NPCMTWEE682 pKa = 3.84QVPIEE687 pKa = 4.05EE688 pKa = 5.28LNFSGYY694 pKa = 10.85NGVGDD699 pKa = 4.14IIYY702 pKa = 10.19RR703 pKa = 11.84EE704 pKa = 4.0NNQTVIINISEE715 pKa = 4.53PFTISQSLNIDD726 pKa = 3.48GLKK729 pKa = 10.94VNICNTNFYY738 pKa = 10.71NGAGLEE744 pKa = 3.98YY745 pKa = 10.32SIRR748 pKa = 11.84SDD750 pKa = 3.12PTFYY754 pKa = 11.05LHH756 pKa = 7.88DD757 pKa = 4.44NGEE760 pKa = 4.72HH761 pKa = 5.87ICKK764 pKa = 9.91SSKK767 pKa = 10.44PIITAIEE774 pKa = 3.96DD775 pKa = 3.69EE776 pKa = 4.56YY777 pKa = 12.0VNINSISEE785 pKa = 4.46MITKK789 pKa = 10.24LIIKK793 pKa = 9.02EE794 pKa = 4.06DD795 pKa = 3.61SVLPIINTTRR805 pKa = 11.84KK806 pKa = 9.0IVIWAPDD813 pKa = 3.81PLIWDD818 pKa = 3.93SNISLVSLEE827 pKa = 4.37GQGKK831 pKa = 7.37TKK833 pKa = 10.46VSPEE837 pKa = 3.76LTYY840 pKa = 11.52SNDD843 pKa = 3.34NKK845 pKa = 10.32QLQLDD850 pKa = 3.76ITDD853 pKa = 3.88SFSAFDD859 pKa = 3.62SLVISGMRR867 pKa = 11.84VKK869 pKa = 10.77KK870 pKa = 10.48DD871 pKa = 3.21GNYY874 pKa = 10.5SGDD877 pKa = 3.78FASSAHH883 pKa = 6.0FKK885 pKa = 10.31MSLPEE890 pKa = 5.04SIDD893 pKa = 3.16SFYY896 pKa = 10.51PIHH899 pKa = 7.17LDD901 pKa = 3.3NSSSDD906 pKa = 3.3LSIIYY911 pKa = 10.35SSAHH915 pKa = 6.28LEE917 pKa = 4.23FEE919 pKa = 4.47SSQGANKK926 pKa = 9.92IVFIKK931 pKa = 10.77GGANSEE937 pKa = 4.35YY938 pKa = 10.72FNVTIFGGSLSNSINIDD955 pKa = 3.16TDD957 pKa = 3.22IKK959 pKa = 11.38LMLPTFLNVHH969 pKa = 4.77WVEE972 pKa = 4.12NQSAQIDD979 pKa = 4.32PGTPAHH985 pKa = 7.32IINNVASINSLAILPNTNNKK1005 pKa = 8.5VLNLNVLEE1013 pKa = 4.63AFSINDD1019 pKa = 3.63SIKK1022 pKa = 10.37INNLSVEE1029 pKa = 4.27NNTTVNDD1036 pKa = 3.71MSKK1039 pKa = 9.94IDD1041 pKa = 4.14YY1042 pKa = 10.73LSLLQGDD1049 pKa = 4.23YY1050 pKa = 11.24GSNNHH1055 pKa = 7.43AINQADD1061 pKa = 4.15SLIVAAPQMMIAKK1074 pKa = 9.56DD1075 pKa = 3.14ITVITGALEE1084 pKa = 4.51NINIPPITIVDD1095 pKa = 3.75DD1096 pKa = 3.78TLEE1099 pKa = 3.93QVLNIGNNNFYY1110 pKa = 10.69YY1111 pKa = 10.78LSLEE1115 pKa = 4.58GISNYY1120 pKa = 9.73EE1121 pKa = 3.57WDD1123 pKa = 4.75SGFTCLNMLCTLSDD1137 pKa = 3.11NKK1139 pKa = 10.07TIKK1142 pKa = 10.45FLIQSSGTLTISNLKK1157 pKa = 9.61LKK1159 pKa = 10.71NVTFDD1164 pKa = 3.61NTDD1167 pKa = 3.11DD1168 pKa = 4.01AYY1170 pKa = 11.43RR1171 pKa = 11.84LVLKK1175 pKa = 10.65DD1176 pKa = 4.17NNTKK1180 pKa = 10.08MIAKK1184 pKa = 9.69SINTIKK1190 pKa = 10.04TGMPKK1195 pKa = 9.13IDD1197 pKa = 5.8LITGVIWVDD1206 pKa = 3.39LDD1208 pKa = 4.0SIKK1211 pKa = 10.82AISTLKK1217 pKa = 10.19IIEE1220 pKa = 4.74SINHH1224 pKa = 6.13ISSDD1228 pKa = 3.7SLKK1231 pKa = 10.91LYY1233 pKa = 10.4FDD1235 pKa = 3.57KK1236 pKa = 11.11SYY1238 pKa = 11.27LKK1240 pKa = 9.84WDD1242 pKa = 3.69QQSSVQSTINYY1253 pKa = 7.23GFPSEE1258 pKa = 4.28GEE1260 pKa = 4.09LYY1262 pKa = 10.41INQYY1266 pKa = 11.0EE1267 pKa = 4.23NGSNSAFDD1275 pKa = 4.27LSAFNFLITSDD1286 pKa = 3.67VFKK1289 pKa = 10.78DD1290 pKa = 2.97ITQADD1295 pKa = 3.57TVMNLNCVLMDD1306 pKa = 4.34EE1307 pKa = 4.78YY1308 pKa = 11.67NQFTITHH1315 pKa = 6.9PIILNFDD1322 pKa = 3.99PLISDD1327 pKa = 4.41VIPLGGTTWDD1337 pKa = 3.65SLTLSVIINEE1347 pKa = 4.21NFLEE1351 pKa = 5.14DD1352 pKa = 3.83EE1353 pKa = 4.44GTDD1356 pKa = 3.53LSSIRR1361 pKa = 11.84PTLLLLDD1368 pKa = 4.49GEE1370 pKa = 5.35GDD1372 pKa = 3.48TSNYY1376 pKa = 10.35DD1377 pKa = 2.86IDD1379 pKa = 4.25VSFTQTAAQGDD1390 pKa = 4.44FPEE1393 pKa = 4.68GVYY1396 pKa = 9.07TAEE1399 pKa = 4.83FYY1401 pKa = 11.06FPDD1404 pKa = 3.7SLIDD1408 pKa = 3.46ILTSKK1413 pKa = 10.68QEE1415 pKa = 4.01NNDD1418 pKa = 2.88IDD1420 pKa = 4.98LLLSLTDD1427 pKa = 3.36IDD1429 pKa = 4.82SNALSRR1435 pKa = 11.84SNSSKK1440 pKa = 11.15DD1441 pKa = 3.12NWKK1444 pKa = 8.89LTPISLGWSEE1454 pKa = 5.87DD1455 pKa = 2.91ITNIYY1460 pKa = 10.01LDD1462 pKa = 3.16QSLRR1466 pKa = 11.84YY1467 pKa = 9.75LNPDD1471 pKa = 3.03SNDD1474 pKa = 2.95IVFTLPEE1481 pKa = 5.16PISTTCEE1488 pKa = 3.23IRR1490 pKa = 11.84INNLVSDD1497 pKa = 4.07STYY1500 pKa = 11.11EE1501 pKa = 4.0LGTMAINEE1509 pKa = 4.15NSTITIEE1516 pKa = 4.13GTQIQNALGRR1526 pKa = 11.84DD1527 pKa = 3.09ADD1529 pKa = 4.41GIYY1532 pKa = 10.34DD1533 pKa = 5.27IILQDD1538 pKa = 3.3KK1539 pKa = 10.53SGGSFSFPFIKK1550 pKa = 10.25RR1551 pKa = 11.84IIIDD1555 pKa = 3.7TSPPQFISINPTTDD1569 pKa = 2.9VFDD1572 pKa = 4.36SGWNEE1577 pKa = 3.81NSVHH1581 pKa = 7.11SITDD1585 pKa = 2.99QDD1587 pKa = 4.63KK1588 pKa = 11.0IILTFKK1594 pKa = 10.14DD1595 pKa = 4.09YY1596 pKa = 11.03PIMTTVDD1603 pKa = 3.58SIFIYY1608 pKa = 10.78NSGIVSAMAPGGYY1621 pKa = 9.88LFNDD1625 pKa = 4.28SIDD1628 pKa = 3.47IQISFEE1634 pKa = 4.16LTDD1637 pKa = 3.66SNGNDD1642 pKa = 3.11TSYY1645 pKa = 10.81TFLNTDD1651 pKa = 3.37EE1652 pKa = 5.41NGLLDD1657 pKa = 4.44FYY1659 pKa = 11.19KK1660 pKa = 10.75EE1661 pKa = 4.31SIIKK1665 pKa = 10.68LSDD1668 pKa = 3.22HH1669 pKa = 5.28QLEE1672 pKa = 4.63GKK1674 pKa = 10.17NSIIINYY1681 pKa = 8.55VLSDD1685 pKa = 3.47HH1686 pKa = 7.26AGNEE1690 pKa = 3.75QNYY1693 pKa = 7.55TIKK1696 pKa = 10.8YY1697 pKa = 8.05VLYY1700 pKa = 9.97NASDD1704 pKa = 3.4IQGLVDD1710 pKa = 6.26DD1711 pKa = 5.25IFNFPNPFSSVDD1723 pKa = 2.79EE1724 pKa = 4.37GTRR1727 pKa = 11.84IRR1729 pKa = 11.84YY1730 pKa = 9.47SLTQDD1735 pKa = 3.23GNQGKK1740 pKa = 9.3YY1741 pKa = 7.46VVFNAKK1747 pKa = 10.4GEE1749 pKa = 4.25LVYY1752 pKa = 10.41HH1753 pKa = 6.19YY1754 pKa = 10.03SLSSSEE1760 pKa = 4.19LQQGTHH1766 pKa = 6.09TIYY1769 pKa = 10.47WDD1771 pKa = 3.28GHH1773 pKa = 4.78YY1774 pKa = 11.08LNNNYY1779 pKa = 10.28LLATGVYY1786 pKa = 10.21FGFLDD1791 pKa = 4.37IDD1793 pKa = 4.11DD1794 pKa = 5.66KK1795 pKa = 11.12IAKK1798 pKa = 9.63HH1799 pKa = 6.23KK1800 pKa = 10.38IAIINN1805 pKa = 3.58
SS1 pKa = 6.79NGGLGGSTVIATVTVNPVNDD21 pKa = 3.86EE22 pKa = 4.03PSFEE26 pKa = 4.28YY27 pKa = 10.92TNVVAVNEE35 pKa = 4.05DD36 pKa = 3.32SGLYY40 pKa = 7.63EE41 pKa = 4.5ASWAFNISVGPDD53 pKa = 3.25NEE55 pKa = 4.47SDD57 pKa = 3.59QIPTFEE63 pKa = 5.38IISNDD68 pKa = 3.2HH69 pKa = 6.33EE70 pKa = 5.51SYY72 pKa = 10.98FSIQPSISEE81 pKa = 4.23LGVLTFTPADD91 pKa = 3.8NQHH94 pKa = 6.04GEE96 pKa = 4.09AIITVRR102 pKa = 11.84LTDD105 pKa = 3.55NGGTEE110 pKa = 3.97NGGVNTYY117 pKa = 11.09DD118 pKa = 3.9DD119 pKa = 3.41QTFTIKK125 pKa = 10.63LNPIADD131 pKa = 4.08TPNITDD137 pKa = 3.81AEE139 pKa = 4.16TDD141 pKa = 3.42EE142 pKa = 6.81DD143 pKa = 4.04IMSSSGLVITRR154 pKa = 11.84NAVDD158 pKa = 3.77SEE160 pKa = 4.52EE161 pKa = 4.12VTHH164 pKa = 7.03FKK166 pKa = 9.2ITGITNGTLYY176 pKa = 10.68QNNGSTSIANNEE188 pKa = 4.26FITFTEE194 pKa = 4.74GNTGLKK200 pKa = 9.9FDD202 pKa = 5.24PDD204 pKa = 3.11EE205 pKa = 5.15DD206 pKa = 5.24YY207 pKa = 11.3YY208 pKa = 11.73GSGFITIQASVSSSNGGLGGSTVIATVTVNPEE240 pKa = 3.41NDD242 pKa = 3.5GPYY245 pKa = 10.26DD246 pKa = 3.8AEE248 pKa = 6.0AGDD251 pKa = 5.2DD252 pKa = 4.07IYY254 pKa = 11.41DD255 pKa = 4.15ILPSDD260 pKa = 3.82ATTIDD265 pKa = 3.28ITILDD270 pKa = 4.11VSSDD274 pKa = 4.41DD275 pKa = 3.66IDD277 pKa = 6.61DD278 pKa = 4.54DD279 pKa = 4.82VSVSHH284 pKa = 6.76TYY286 pKa = 10.1SWTLIDD292 pKa = 5.1ANNGDD297 pKa = 4.61IIAEE301 pKa = 3.98ATEE304 pKa = 4.06INPTFALKK312 pKa = 10.39AGHH315 pKa = 5.79YY316 pKa = 8.9QFEE319 pKa = 4.7LTVTDD324 pKa = 3.49TTTNDD329 pKa = 2.94SNIYY333 pKa = 10.86GDD335 pKa = 3.52ITDD338 pKa = 4.11TDD340 pKa = 4.02TIDD343 pKa = 3.76IYY345 pKa = 10.68IGQPGVDD352 pKa = 3.08ISSNPVFVTYY362 pKa = 11.05DD363 pKa = 3.28DD364 pKa = 3.34VAYY367 pKa = 9.94IDD369 pKa = 3.32IVYY372 pKa = 10.27FEE374 pKa = 5.05HH375 pKa = 7.44EE376 pKa = 4.54YY377 pKa = 11.46ASVTQNGDD385 pKa = 3.37QIVISVPIEE394 pKa = 3.97YY395 pKa = 10.11QNKK398 pKa = 7.66FQFIAGQIIPQDD410 pKa = 3.42ALEE413 pKa = 4.08IQIGYY418 pKa = 9.65SGDD421 pKa = 3.42VTDD424 pKa = 5.71ISIEE428 pKa = 4.32SITTSAITINVVTSHH443 pKa = 6.41ISEE446 pKa = 3.99NFYY449 pKa = 10.82IKK451 pKa = 10.57LKK453 pKa = 10.19SVEE456 pKa = 4.19VNPIDD461 pKa = 3.74VTSDD465 pKa = 3.41FSLEE469 pKa = 4.06IEE471 pKa = 4.22NDD473 pKa = 3.03NYY475 pKa = 11.46LPILKK480 pKa = 9.91SATEE484 pKa = 3.82ISIGNPKK491 pKa = 8.31ITTITDD497 pKa = 3.35QVLITANPNEE507 pKa = 4.23KK508 pKa = 9.44MLDD511 pKa = 3.35IIVYY515 pKa = 9.56EE516 pKa = 4.46DD517 pKa = 3.7EE518 pKa = 4.34SVAAINTDD526 pKa = 3.61DD527 pKa = 5.84DD528 pKa = 3.95IVIQFPSTLEE538 pKa = 4.15NNVKK542 pKa = 9.8WIQVPTASSNSEE554 pKa = 4.1KK555 pKa = 10.49IGTITIDD562 pKa = 3.38SSDD565 pKa = 3.77STKK568 pKa = 11.16LLIDD572 pKa = 4.36VISDD576 pKa = 3.35FFAGEE581 pKa = 4.18SLILTDD587 pKa = 5.28GYY589 pKa = 11.02VQVGDD594 pKa = 3.89IEE596 pKa = 5.24SNEE599 pKa = 3.71DD600 pKa = 3.26DD601 pKa = 4.07YY602 pKa = 12.12YY603 pKa = 11.59KK604 pKa = 10.54IIAYY608 pKa = 10.21VNGRR612 pKa = 11.84NQSLDD617 pKa = 3.61VEE619 pKa = 4.76CDD621 pKa = 2.8NSIIVTSPEE630 pKa = 3.59LSFASDD636 pKa = 2.57QHH638 pKa = 6.88YY639 pKa = 10.64VVNDD643 pKa = 3.61SAVSFPTITLTEE655 pKa = 4.1SSLIPSIYY663 pKa = 10.53SEE665 pKa = 4.35VQDD668 pKa = 4.23SIYY671 pKa = 10.84LRR673 pKa = 11.84FDD675 pKa = 3.68NPCMTWEE682 pKa = 3.84QVPIEE687 pKa = 4.05EE688 pKa = 5.28LNFSGYY694 pKa = 10.85NGVGDD699 pKa = 4.14IIYY702 pKa = 10.19RR703 pKa = 11.84EE704 pKa = 4.0NNQTVIINISEE715 pKa = 4.53PFTISQSLNIDD726 pKa = 3.48GLKK729 pKa = 10.94VNICNTNFYY738 pKa = 10.71NGAGLEE744 pKa = 3.98YY745 pKa = 10.32SIRR748 pKa = 11.84SDD750 pKa = 3.12PTFYY754 pKa = 11.05LHH756 pKa = 7.88DD757 pKa = 4.44NGEE760 pKa = 4.72HH761 pKa = 5.87ICKK764 pKa = 9.91SSKK767 pKa = 10.44PIITAIEE774 pKa = 3.96DD775 pKa = 3.69EE776 pKa = 4.56YY777 pKa = 12.0VNINSISEE785 pKa = 4.46MITKK789 pKa = 10.24LIIKK793 pKa = 9.02EE794 pKa = 4.06DD795 pKa = 3.61SVLPIINTTRR805 pKa = 11.84KK806 pKa = 9.0IVIWAPDD813 pKa = 3.81PLIWDD818 pKa = 3.93SNISLVSLEE827 pKa = 4.37GQGKK831 pKa = 7.37TKK833 pKa = 10.46VSPEE837 pKa = 3.76LTYY840 pKa = 11.52SNDD843 pKa = 3.34NKK845 pKa = 10.32QLQLDD850 pKa = 3.76ITDD853 pKa = 3.88SFSAFDD859 pKa = 3.62SLVISGMRR867 pKa = 11.84VKK869 pKa = 10.77KK870 pKa = 10.48DD871 pKa = 3.21GNYY874 pKa = 10.5SGDD877 pKa = 3.78FASSAHH883 pKa = 6.0FKK885 pKa = 10.31MSLPEE890 pKa = 5.04SIDD893 pKa = 3.16SFYY896 pKa = 10.51PIHH899 pKa = 7.17LDD901 pKa = 3.3NSSSDD906 pKa = 3.3LSIIYY911 pKa = 10.35SSAHH915 pKa = 6.28LEE917 pKa = 4.23FEE919 pKa = 4.47SSQGANKK926 pKa = 9.92IVFIKK931 pKa = 10.77GGANSEE937 pKa = 4.35YY938 pKa = 10.72FNVTIFGGSLSNSINIDD955 pKa = 3.16TDD957 pKa = 3.22IKK959 pKa = 11.38LMLPTFLNVHH969 pKa = 4.77WVEE972 pKa = 4.12NQSAQIDD979 pKa = 4.32PGTPAHH985 pKa = 7.32IINNVASINSLAILPNTNNKK1005 pKa = 8.5VLNLNVLEE1013 pKa = 4.63AFSINDD1019 pKa = 3.63SIKK1022 pKa = 10.37INNLSVEE1029 pKa = 4.27NNTTVNDD1036 pKa = 3.71MSKK1039 pKa = 9.94IDD1041 pKa = 4.14YY1042 pKa = 10.73LSLLQGDD1049 pKa = 4.23YY1050 pKa = 11.24GSNNHH1055 pKa = 7.43AINQADD1061 pKa = 4.15SLIVAAPQMMIAKK1074 pKa = 9.56DD1075 pKa = 3.14ITVITGALEE1084 pKa = 4.51NINIPPITIVDD1095 pKa = 3.75DD1096 pKa = 3.78TLEE1099 pKa = 3.93QVLNIGNNNFYY1110 pKa = 10.69YY1111 pKa = 10.78LSLEE1115 pKa = 4.58GISNYY1120 pKa = 9.73EE1121 pKa = 3.57WDD1123 pKa = 4.75SGFTCLNMLCTLSDD1137 pKa = 3.11NKK1139 pKa = 10.07TIKK1142 pKa = 10.45FLIQSSGTLTISNLKK1157 pKa = 9.61LKK1159 pKa = 10.71NVTFDD1164 pKa = 3.61NTDD1167 pKa = 3.11DD1168 pKa = 4.01AYY1170 pKa = 11.43RR1171 pKa = 11.84LVLKK1175 pKa = 10.65DD1176 pKa = 4.17NNTKK1180 pKa = 10.08MIAKK1184 pKa = 9.69SINTIKK1190 pKa = 10.04TGMPKK1195 pKa = 9.13IDD1197 pKa = 5.8LITGVIWVDD1206 pKa = 3.39LDD1208 pKa = 4.0SIKK1211 pKa = 10.82AISTLKK1217 pKa = 10.19IIEE1220 pKa = 4.74SINHH1224 pKa = 6.13ISSDD1228 pKa = 3.7SLKK1231 pKa = 10.91LYY1233 pKa = 10.4FDD1235 pKa = 3.57KK1236 pKa = 11.11SYY1238 pKa = 11.27LKK1240 pKa = 9.84WDD1242 pKa = 3.69QQSSVQSTINYY1253 pKa = 7.23GFPSEE1258 pKa = 4.28GEE1260 pKa = 4.09LYY1262 pKa = 10.41INQYY1266 pKa = 11.0EE1267 pKa = 4.23NGSNSAFDD1275 pKa = 4.27LSAFNFLITSDD1286 pKa = 3.67VFKK1289 pKa = 10.78DD1290 pKa = 2.97ITQADD1295 pKa = 3.57TVMNLNCVLMDD1306 pKa = 4.34EE1307 pKa = 4.78YY1308 pKa = 11.67NQFTITHH1315 pKa = 6.9PIILNFDD1322 pKa = 3.99PLISDD1327 pKa = 4.41VIPLGGTTWDD1337 pKa = 3.65SLTLSVIINEE1347 pKa = 4.21NFLEE1351 pKa = 5.14DD1352 pKa = 3.83EE1353 pKa = 4.44GTDD1356 pKa = 3.53LSSIRR1361 pKa = 11.84PTLLLLDD1368 pKa = 4.49GEE1370 pKa = 5.35GDD1372 pKa = 3.48TSNYY1376 pKa = 10.35DD1377 pKa = 2.86IDD1379 pKa = 4.25VSFTQTAAQGDD1390 pKa = 4.44FPEE1393 pKa = 4.68GVYY1396 pKa = 9.07TAEE1399 pKa = 4.83FYY1401 pKa = 11.06FPDD1404 pKa = 3.7SLIDD1408 pKa = 3.46ILTSKK1413 pKa = 10.68QEE1415 pKa = 4.01NNDD1418 pKa = 2.88IDD1420 pKa = 4.98LLLSLTDD1427 pKa = 3.36IDD1429 pKa = 4.82SNALSRR1435 pKa = 11.84SNSSKK1440 pKa = 11.15DD1441 pKa = 3.12NWKK1444 pKa = 8.89LTPISLGWSEE1454 pKa = 5.87DD1455 pKa = 2.91ITNIYY1460 pKa = 10.01LDD1462 pKa = 3.16QSLRR1466 pKa = 11.84YY1467 pKa = 9.75LNPDD1471 pKa = 3.03SNDD1474 pKa = 2.95IVFTLPEE1481 pKa = 5.16PISTTCEE1488 pKa = 3.23IRR1490 pKa = 11.84INNLVSDD1497 pKa = 4.07STYY1500 pKa = 11.11EE1501 pKa = 4.0LGTMAINEE1509 pKa = 4.15NSTITIEE1516 pKa = 4.13GTQIQNALGRR1526 pKa = 11.84DD1527 pKa = 3.09ADD1529 pKa = 4.41GIYY1532 pKa = 10.34DD1533 pKa = 5.27IILQDD1538 pKa = 3.3KK1539 pKa = 10.53SGGSFSFPFIKK1550 pKa = 10.25RR1551 pKa = 11.84IIIDD1555 pKa = 3.7TSPPQFISINPTTDD1569 pKa = 2.9VFDD1572 pKa = 4.36SGWNEE1577 pKa = 3.81NSVHH1581 pKa = 7.11SITDD1585 pKa = 2.99QDD1587 pKa = 4.63KK1588 pKa = 11.0IILTFKK1594 pKa = 10.14DD1595 pKa = 4.09YY1596 pKa = 11.03PIMTTVDD1603 pKa = 3.58SIFIYY1608 pKa = 10.78NSGIVSAMAPGGYY1621 pKa = 9.88LFNDD1625 pKa = 4.28SIDD1628 pKa = 3.47IQISFEE1634 pKa = 4.16LTDD1637 pKa = 3.66SNGNDD1642 pKa = 3.11TSYY1645 pKa = 10.81TFLNTDD1651 pKa = 3.37EE1652 pKa = 5.41NGLLDD1657 pKa = 4.44FYY1659 pKa = 11.19KK1660 pKa = 10.75EE1661 pKa = 4.31SIIKK1665 pKa = 10.68LSDD1668 pKa = 3.22HH1669 pKa = 5.28QLEE1672 pKa = 4.63GKK1674 pKa = 10.17NSIIINYY1681 pKa = 8.55VLSDD1685 pKa = 3.47HH1686 pKa = 7.26AGNEE1690 pKa = 3.75QNYY1693 pKa = 7.55TIKK1696 pKa = 10.8YY1697 pKa = 8.05VLYY1700 pKa = 9.97NASDD1704 pKa = 3.4IQGLVDD1710 pKa = 6.26DD1711 pKa = 5.25IFNFPNPFSSVDD1723 pKa = 2.79EE1724 pKa = 4.37GTRR1727 pKa = 11.84IRR1729 pKa = 11.84YY1730 pKa = 9.47SLTQDD1735 pKa = 3.23GNQGKK1740 pKa = 9.3YY1741 pKa = 7.46VVFNAKK1747 pKa = 10.4GEE1749 pKa = 4.25LVYY1752 pKa = 10.41HH1753 pKa = 6.19YY1754 pKa = 10.03SLSSSEE1760 pKa = 4.19LQQGTHH1766 pKa = 6.09TIYY1769 pKa = 10.47WDD1771 pKa = 3.28GHH1773 pKa = 4.78YY1774 pKa = 11.08LNNNYY1779 pKa = 10.28LLATGVYY1786 pKa = 10.21FGFLDD1791 pKa = 4.37IDD1793 pKa = 4.11DD1794 pKa = 5.66KK1795 pKa = 11.12IAKK1798 pKa = 9.63HH1799 pKa = 6.23KK1800 pKa = 10.38IAIINN1805 pKa = 3.58
Molecular weight: 199.05 kDa
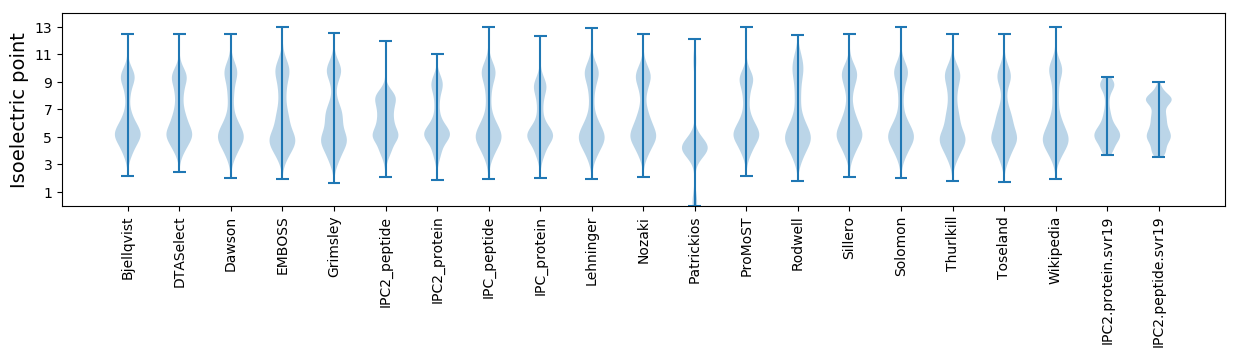
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3M7KIZ1|A0A3M7KIZ1_9BACT Electron transfer flavoprotein-ubiquinone oxidoreductase OS=Candidatus Marinimicrobia bacterium PRS2 OX=2483458 GN=EB821_05175 PE=4 SV=1
MM1 pKa = 7.53KK2 pKa = 9.93VTIKK6 pKa = 9.38YY7 pKa = 7.53TNRR10 pKa = 11.84KK11 pKa = 8.69RR12 pKa = 11.84KK13 pKa = 9.28NKK15 pKa = 10.0HH16 pKa = 4.06GFKK19 pKa = 10.59KK20 pKa = 10.51RR21 pKa = 11.84MSTKK25 pKa = 10.33AGRR28 pKa = 11.84ALLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.79KK37 pKa = 10.3GRR39 pKa = 11.84KK40 pKa = 7.34TISAA44 pKa = 3.98
MM1 pKa = 7.53KK2 pKa = 9.93VTIKK6 pKa = 9.38YY7 pKa = 7.53TNRR10 pKa = 11.84KK11 pKa = 8.69RR12 pKa = 11.84KK13 pKa = 9.28NKK15 pKa = 10.0HH16 pKa = 4.06GFKK19 pKa = 10.59KK20 pKa = 10.51RR21 pKa = 11.84MSTKK25 pKa = 10.33AGRR28 pKa = 11.84ALLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.79KK37 pKa = 10.3GRR39 pKa = 11.84KK40 pKa = 7.34TISAA44 pKa = 3.98
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
424383 |
35 |
2954 |
351.0 |
39.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.624 ± 0.073 | 1.382 ± 0.101 |
6.442 ± 0.1 | 6.719 ± 0.069 |
4.701 ± 0.063 | 7.495 ± 0.097 |
1.892 ± 0.032 | 8.633 ± 0.082 |
5.982 ± 0.113 | 8.976 ± 0.082 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.344 ± 0.034 | 6.087 ± 0.082 |
3.822 ± 0.035 | 3.179 ± 0.046 |
3.414 ± 0.059 | 7.087 ± 0.068 |
5.334 ± 0.043 | 5.649 ± 0.054 |
1.301 ± 0.03 | 3.937 ± 0.053 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
