
Bacillus sp. TS-2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; unclassified Bacillus (in: Bacteria)
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
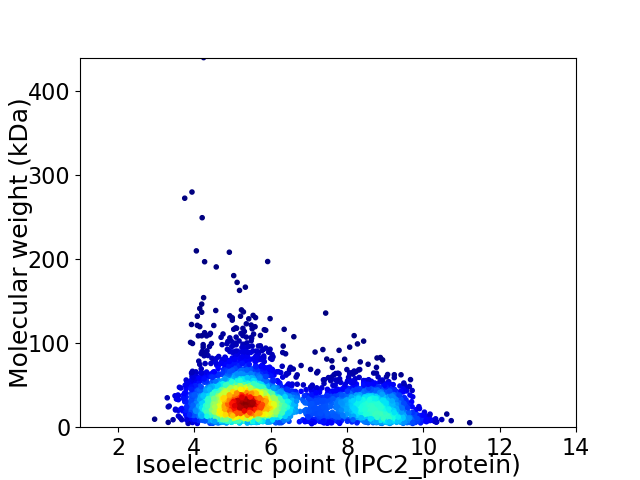
Virtual 2D-PAGE plot for 4084 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|X0RAK5|X0RAK5_9BACI Putative alcohol dehydrogenase OS=Bacillus sp. TS-2 OX=1450694 GN=BTS2_2949 PE=3 SV=1
MM1 pKa = 7.62KK2 pKa = 10.54KK3 pKa = 10.18MMMACFSICLLFIVVACSSGNSDD26 pKa = 3.28TTSNEE31 pKa = 3.34GSTGDD36 pKa = 4.76QEE38 pKa = 5.89DD39 pKa = 4.5EE40 pKa = 4.3NQSSSVDD47 pKa = 4.14EE48 pKa = 3.94ITIWAWDD55 pKa = 3.79PNFNIAALEE64 pKa = 4.05LAKK67 pKa = 9.97DD68 pKa = 4.05TYY70 pKa = 11.47AEE72 pKa = 4.05EE73 pKa = 4.2NADD76 pKa = 3.47VNINIVEE83 pKa = 4.29SAQDD87 pKa = 4.67DD88 pKa = 4.07IIQRR92 pKa = 11.84LNTILSSGSTSGLPHH107 pKa = 6.48IVLIEE112 pKa = 4.29DD113 pKa = 3.33YY114 pKa = 10.05RR115 pKa = 11.84AQGFLQSYY123 pKa = 8.73PDD125 pKa = 3.83SFHH128 pKa = 7.41EE129 pKa = 3.81ITGAYY134 pKa = 9.42EE135 pKa = 4.32LDD137 pKa = 3.2NFAPYY142 pKa = 10.07KK143 pKa = 10.01IAPTSIDD150 pKa = 3.42DD151 pKa = 4.04ANYY154 pKa = 10.68GLPFDD159 pKa = 4.04TGVAGLYY166 pKa = 10.3VRR168 pKa = 11.84TDD170 pKa = 3.44YY171 pKa = 11.31FEE173 pKa = 4.04EE174 pKa = 5.47AGYY177 pKa = 10.45SVEE180 pKa = 5.39DD181 pKa = 4.13LQSLDD186 pKa = 3.07WNEE189 pKa = 4.17FIEE192 pKa = 4.21IGKK195 pKa = 9.59KK196 pKa = 9.7IKK198 pKa = 10.21EE199 pKa = 4.1ATGKK203 pKa = 10.46YY204 pKa = 10.46LLTLEE209 pKa = 4.83PNNLSQLRR217 pKa = 11.84MMIQTNGGWYY227 pKa = 9.79VEE229 pKa = 4.3EE230 pKa = 5.84DD231 pKa = 3.69GVTPYY236 pKa = 11.0LADD239 pKa = 4.01NEE241 pKa = 4.18DD242 pKa = 3.44LKK244 pKa = 11.13IAFEE248 pKa = 4.39VYY250 pKa = 8.43KK251 pKa = 10.28TLIDD255 pKa = 3.74EE256 pKa = 5.29DD257 pKa = 4.13LAKK260 pKa = 10.47PISDD264 pKa = 3.12WSQFVAGFNSGEE276 pKa = 4.28VASVPTGNWITPSVTAEE293 pKa = 3.94EE294 pKa = 4.46SQSGDD299 pKa = 2.87WAVVPFPTLPGVEE312 pKa = 4.18SSIPATNLGGSSWYY326 pKa = 10.17VLNIDD331 pKa = 4.07GKK333 pKa = 11.04EE334 pKa = 3.93DD335 pKa = 3.46ATDD338 pKa = 4.32FLAKK342 pKa = 9.98TFGSNDD348 pKa = 3.23QLYY351 pKa = 10.44EE352 pKa = 4.24DD353 pKa = 4.95LLNEE357 pKa = 3.94IGAIGTYY364 pKa = 10.46LPASANEE371 pKa = 4.24AYY373 pKa = 10.16QSEE376 pKa = 4.83VEE378 pKa = 4.24FFGNQSIYY386 pKa = 11.27ADD388 pKa = 3.56FAKK391 pKa = 9.94WSEE394 pKa = 4.91LIPQVNYY401 pKa = 10.76GIHH404 pKa = 6.26TYY406 pKa = 10.28SIEE409 pKa = 5.05DD410 pKa = 3.38ILVVEE415 pKa = 4.28MQNYY419 pKa = 10.28LNGKK423 pKa = 9.82DD424 pKa = 3.56IDD426 pKa = 3.95QALQDD431 pKa = 3.83AQVQAEE437 pKa = 4.27TQIRR441 pKa = 3.63
MM1 pKa = 7.62KK2 pKa = 10.54KK3 pKa = 10.18MMMACFSICLLFIVVACSSGNSDD26 pKa = 3.28TTSNEE31 pKa = 3.34GSTGDD36 pKa = 4.76QEE38 pKa = 5.89DD39 pKa = 4.5EE40 pKa = 4.3NQSSSVDD47 pKa = 4.14EE48 pKa = 3.94ITIWAWDD55 pKa = 3.79PNFNIAALEE64 pKa = 4.05LAKK67 pKa = 9.97DD68 pKa = 4.05TYY70 pKa = 11.47AEE72 pKa = 4.05EE73 pKa = 4.2NADD76 pKa = 3.47VNINIVEE83 pKa = 4.29SAQDD87 pKa = 4.67DD88 pKa = 4.07IIQRR92 pKa = 11.84LNTILSSGSTSGLPHH107 pKa = 6.48IVLIEE112 pKa = 4.29DD113 pKa = 3.33YY114 pKa = 10.05RR115 pKa = 11.84AQGFLQSYY123 pKa = 8.73PDD125 pKa = 3.83SFHH128 pKa = 7.41EE129 pKa = 3.81ITGAYY134 pKa = 9.42EE135 pKa = 4.32LDD137 pKa = 3.2NFAPYY142 pKa = 10.07KK143 pKa = 10.01IAPTSIDD150 pKa = 3.42DD151 pKa = 4.04ANYY154 pKa = 10.68GLPFDD159 pKa = 4.04TGVAGLYY166 pKa = 10.3VRR168 pKa = 11.84TDD170 pKa = 3.44YY171 pKa = 11.31FEE173 pKa = 4.04EE174 pKa = 5.47AGYY177 pKa = 10.45SVEE180 pKa = 5.39DD181 pKa = 4.13LQSLDD186 pKa = 3.07WNEE189 pKa = 4.17FIEE192 pKa = 4.21IGKK195 pKa = 9.59KK196 pKa = 9.7IKK198 pKa = 10.21EE199 pKa = 4.1ATGKK203 pKa = 10.46YY204 pKa = 10.46LLTLEE209 pKa = 4.83PNNLSQLRR217 pKa = 11.84MMIQTNGGWYY227 pKa = 9.79VEE229 pKa = 4.3EE230 pKa = 5.84DD231 pKa = 3.69GVTPYY236 pKa = 11.0LADD239 pKa = 4.01NEE241 pKa = 4.18DD242 pKa = 3.44LKK244 pKa = 11.13IAFEE248 pKa = 4.39VYY250 pKa = 8.43KK251 pKa = 10.28TLIDD255 pKa = 3.74EE256 pKa = 5.29DD257 pKa = 4.13LAKK260 pKa = 10.47PISDD264 pKa = 3.12WSQFVAGFNSGEE276 pKa = 4.28VASVPTGNWITPSVTAEE293 pKa = 3.94EE294 pKa = 4.46SQSGDD299 pKa = 2.87WAVVPFPTLPGVEE312 pKa = 4.18SSIPATNLGGSSWYY326 pKa = 10.17VLNIDD331 pKa = 4.07GKK333 pKa = 11.04EE334 pKa = 3.93DD335 pKa = 3.46ATDD338 pKa = 4.32FLAKK342 pKa = 9.98TFGSNDD348 pKa = 3.23QLYY351 pKa = 10.44EE352 pKa = 4.24DD353 pKa = 4.95LLNEE357 pKa = 3.94IGAIGTYY364 pKa = 10.46LPASANEE371 pKa = 4.24AYY373 pKa = 10.16QSEE376 pKa = 4.83VEE378 pKa = 4.24FFGNQSIYY386 pKa = 11.27ADD388 pKa = 3.56FAKK391 pKa = 9.94WSEE394 pKa = 4.91LIPQVNYY401 pKa = 10.76GIHH404 pKa = 6.26TYY406 pKa = 10.28SIEE409 pKa = 5.05DD410 pKa = 3.38ILVVEE415 pKa = 4.28MQNYY419 pKa = 10.28LNGKK423 pKa = 9.82DD424 pKa = 3.56IDD426 pKa = 3.95QALQDD431 pKa = 3.83AQVQAEE437 pKa = 4.27TQIRR441 pKa = 3.63
Molecular weight: 48.75 kDa
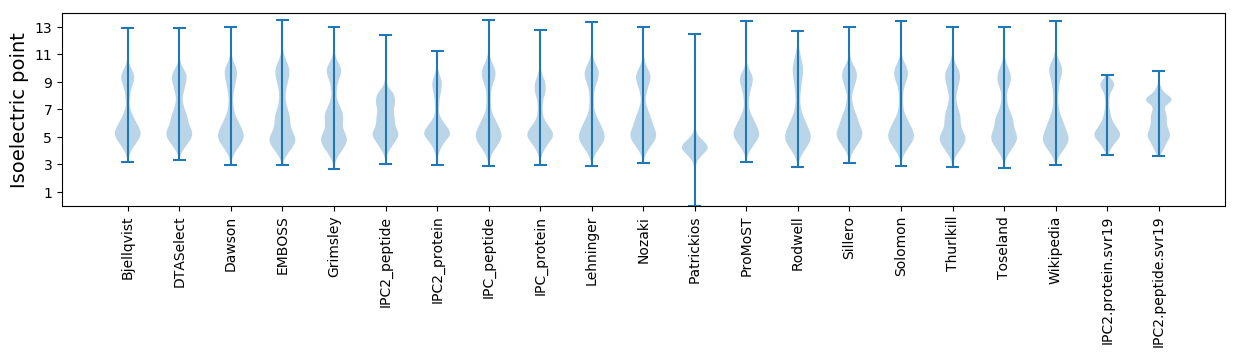
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|X0SRG3|X0SRG3_9BACI Putatice ATPase OS=Bacillus sp. TS-2 OX=1450694 GN=BTS2_3321 PE=4 SV=1
MM1 pKa = 7.71GKK3 pKa = 8.0PTFQPNNRR11 pKa = 11.84KK12 pKa = 9.23RR13 pKa = 11.84KK14 pKa = 8.22KK15 pKa = 8.69VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 10.19NGRR29 pKa = 11.84KK30 pKa = 8.49VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
MM1 pKa = 7.71GKK3 pKa = 8.0PTFQPNNRR11 pKa = 11.84KK12 pKa = 9.23RR13 pKa = 11.84KK14 pKa = 8.22KK15 pKa = 8.69VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 10.19NGRR29 pKa = 11.84KK30 pKa = 8.49VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1205149 |
39 |
3910 |
295.1 |
33.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.154 ± 0.036 | 0.636 ± 0.01 |
5.06 ± 0.028 | 8.138 ± 0.044 |
4.698 ± 0.033 | 6.334 ± 0.039 |
2.226 ± 0.019 | 8.172 ± 0.032 |
6.723 ± 0.047 | 10.009 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.691 ± 0.02 | 4.745 ± 0.033 |
3.419 ± 0.022 | 4.149 ± 0.027 |
3.812 ± 0.025 | 6.46 ± 0.029 |
5.256 ± 0.027 | 6.565 ± 0.029 |
1.088 ± 0.015 | 3.665 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
