
candidate division MSBL1 archaeon SCGC-AAA382K21
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Euryarchaeota incertae sedis; candidate division MSBL1
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
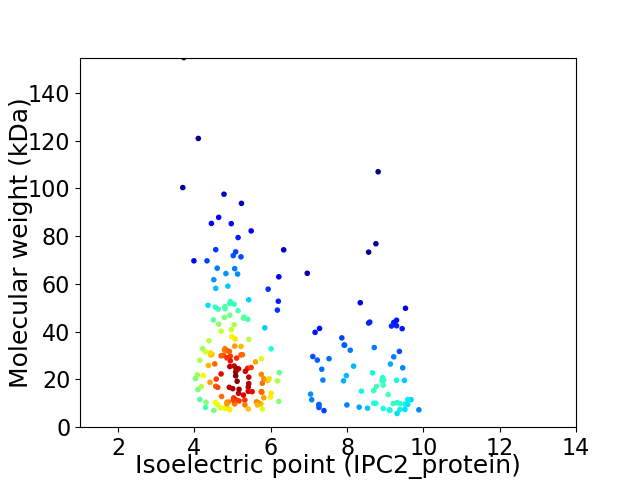
Virtual 2D-PAGE plot for 218 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A133VKM0|A0A133VKM0_9EURY Phosphohydrolase OS=candidate division MSBL1 archaeon SCGC-AAA382K21 OX=1698283 GN=AKJ54_00890 PE=4 SV=1
MM1 pKa = 7.34TWDD4 pKa = 3.31QGNYY8 pKa = 9.41IYY10 pKa = 10.51TLFGGSYY17 pKa = 10.32SDD19 pKa = 3.39TSGDD23 pKa = 3.26SRR25 pKa = 11.84HH26 pKa = 5.53YY27 pKa = 9.55FWRR30 pKa = 11.84YY31 pKa = 10.18DD32 pKa = 3.24ISSDD36 pKa = 3.33SWEE39 pKa = 4.02QLEE42 pKa = 4.5NTPSGQGAGDD52 pKa = 5.16AITWVPGSAVGTSNDD67 pKa = 2.77WVYY70 pKa = 11.33AIVGSKK76 pKa = 10.32VSEE79 pKa = 4.07HH80 pKa = 6.62GIAFYY85 pKa = 10.45RR86 pKa = 11.84YY87 pKa = 10.1NINTNNWQTLQYY99 pKa = 9.11NPSWATEE106 pKa = 3.96GSDD109 pKa = 4.77DD110 pKa = 4.63GSSLVWAGGDD120 pKa = 3.48YY121 pKa = 10.66LYY123 pKa = 11.2SLQGEE128 pKa = 4.44HH129 pKa = 7.07AEE131 pKa = 4.7SDD133 pKa = 3.87PADD136 pKa = 3.64DD137 pKa = 3.71RR138 pKa = 11.84AFARR142 pKa = 11.84FDD144 pKa = 3.13ITDD147 pKa = 3.74NSWTDD152 pKa = 3.25LDD154 pKa = 5.57DD155 pKa = 4.01IPEE158 pKa = 4.3EE159 pKa = 4.47GGVGDD164 pKa = 4.43GGSLAWAGGNYY175 pKa = 9.37SNKK178 pKa = 9.46VYY180 pKa = 10.9ALGGGFVGRR189 pKa = 11.84GEE191 pKa = 4.27TPGDD195 pKa = 3.15NFYY198 pKa = 11.4VYY200 pKa = 9.17DD201 pKa = 3.89TSVDD205 pKa = 2.81NWTQLQSLPYY215 pKa = 10.89GMTGQNGPRR224 pKa = 11.84LGYY227 pKa = 10.8ADD229 pKa = 3.67GSLYY233 pKa = 10.4CWRR236 pKa = 11.84GYY238 pKa = 11.12YY239 pKa = 9.86DD240 pKa = 3.97ASNNPDD246 pKa = 2.84VLMAYY251 pKa = 7.54EE252 pKa = 4.53LPAVGVSVSISPSEE266 pKa = 3.88NNALNGEE273 pKa = 4.34NVTFTVTVTNVDD285 pKa = 3.98DD286 pKa = 3.74EE287 pKa = 4.48HH288 pKa = 8.42HH289 pKa = 6.6NFVLEE294 pKa = 4.27NSDD297 pKa = 3.69SLDD300 pKa = 3.21WQMSLSEE307 pKa = 5.35DD308 pKa = 3.91NLSLDD313 pKa = 3.97PDD315 pKa = 3.41NSEE318 pKa = 4.04NVVLEE323 pKa = 4.03VAIPPAVSGGTEE335 pKa = 3.93DD336 pKa = 3.8TVTVTATAQDD346 pKa = 3.49NAEE349 pKa = 4.13ISSSAGCTAGAQVDD363 pKa = 3.45VDD365 pKa = 4.1FNVWITPFSQTGSRR379 pKa = 11.84EE380 pKa = 3.96DD381 pKa = 3.35NLDD384 pKa = 3.54YY385 pKa = 11.0VVHH388 pKa = 6.27VKK390 pKa = 10.85NSGNVVDD397 pKa = 5.59NYY399 pKa = 11.7DD400 pKa = 5.98LIITDD405 pKa = 3.56NAGWADD411 pKa = 3.78NVSLAGSGYY420 pKa = 10.66SDD422 pKa = 4.52ADD424 pKa = 3.86NIFPSADD431 pKa = 3.16AGICEE436 pKa = 4.69YY437 pKa = 11.14YY438 pKa = 10.34PNTTKK443 pKa = 10.99GGDD446 pKa = 3.3NYY448 pKa = 11.02NVYY451 pKa = 10.41VGYY454 pKa = 10.52EE455 pKa = 3.91NNPYY459 pKa = 10.29KK460 pKa = 10.38PNCTGEE466 pKa = 3.98DD467 pKa = 3.15TRR469 pKa = 11.84ALFKK473 pKa = 10.67FDD475 pKa = 4.8LSGIPDD481 pKa = 4.13GATITGATLNAHH493 pKa = 6.75GDD495 pKa = 4.09FSPSTGFPNYY505 pKa = 9.6NDD507 pKa = 4.89RR508 pKa = 11.84IMLTDD513 pKa = 3.65VQPLGSDD520 pKa = 2.6SWGEE524 pKa = 3.81NTVNWNNKK532 pKa = 8.55PSFGSTLDD540 pKa = 3.49SVLIDD545 pKa = 4.66DD546 pKa = 4.67NEE548 pKa = 4.06YY549 pKa = 10.34WSWNVKK555 pKa = 10.11SAVQNEE561 pKa = 4.09VSGDD565 pKa = 3.75GVVSLGLLSEE575 pKa = 4.36NAVSNQKK582 pKa = 10.13NVTMWFHH589 pKa = 6.28TKK591 pKa = 10.32DD592 pKa = 3.69GSPEE596 pKa = 3.93NPDD599 pKa = 3.14SVKK602 pKa = 10.47PYY604 pKa = 11.02LEE606 pKa = 4.03VLYY609 pKa = 10.15EE610 pKa = 4.18VYY612 pKa = 10.74DD613 pKa = 3.91VVSLKK618 pKa = 10.84NISPGEE624 pKa = 4.06NEE626 pKa = 3.84NVALNVTIPNSAASGDD642 pKa = 3.77HH643 pKa = 4.73TVKK646 pKa = 10.85VGATSYY652 pKa = 11.78GDD654 pKa = 3.8DD655 pKa = 3.95SLSKK659 pKa = 10.46EE660 pKa = 4.22NSCIASVGSPTGVGVSISPSEE681 pKa = 4.58DD682 pKa = 3.18NAPPGYY688 pKa = 9.89GLNYY692 pKa = 9.84EE693 pKa = 4.33VTVTNTGSISDD704 pKa = 4.19NYY706 pKa = 10.62DD707 pKa = 3.13LSVVDD712 pKa = 4.1NAVPSWNPTLDD723 pKa = 3.14NSLFEE728 pKa = 4.75DD729 pKa = 4.07VPPGEE734 pKa = 4.19NRR736 pKa = 11.84NTVLRR741 pKa = 11.84VTVPEE746 pKa = 3.91NANLGVEE753 pKa = 4.28DD754 pKa = 4.82NIIVTASSQEE764 pKa = 4.02NVEE767 pKa = 3.93MDD769 pKa = 4.53DD770 pKa = 3.76NASCIALSAALFRR783 pKa = 11.84SVKK786 pKa = 10.11ISVSPSYY793 pKa = 10.62KK794 pKa = 10.11AGSSGSSLDD803 pKa = 3.49YY804 pKa = 10.67SVKK807 pKa = 10.26VKK809 pKa = 10.72NRR811 pKa = 11.84GNIPDD816 pKa = 3.88NYY818 pKa = 9.85EE819 pKa = 4.04LEE821 pKa = 4.65ASDD824 pKa = 5.82DD825 pKa = 4.21VFPSWDD831 pKa = 3.45PTVVDD836 pKa = 4.58NIFLNVVPEE845 pKa = 4.17EE846 pKa = 4.16EE847 pKa = 4.38RR848 pKa = 11.84VTTLNVVIPEE858 pKa = 3.96NDD860 pKa = 3.22NLRR863 pKa = 11.84GIEE866 pKa = 4.08DD867 pKa = 4.47NITTTVTSLEE877 pKa = 4.11NDD879 pKa = 3.78SVHH882 pKa = 7.54DD883 pKa = 4.05SAVCTAQAMDD893 pKa = 5.02SISMQLDD900 pKa = 4.07LVADD904 pKa = 4.13WNLVGLPVVSDD915 pKa = 3.49STTKK919 pKa = 10.58EE920 pKa = 3.69NLFEE924 pKa = 4.3GTGVDD929 pKa = 3.75PLSVV933 pKa = 3.14
MM1 pKa = 7.34TWDD4 pKa = 3.31QGNYY8 pKa = 9.41IYY10 pKa = 10.51TLFGGSYY17 pKa = 10.32SDD19 pKa = 3.39TSGDD23 pKa = 3.26SRR25 pKa = 11.84HH26 pKa = 5.53YY27 pKa = 9.55FWRR30 pKa = 11.84YY31 pKa = 10.18DD32 pKa = 3.24ISSDD36 pKa = 3.33SWEE39 pKa = 4.02QLEE42 pKa = 4.5NTPSGQGAGDD52 pKa = 5.16AITWVPGSAVGTSNDD67 pKa = 2.77WVYY70 pKa = 11.33AIVGSKK76 pKa = 10.32VSEE79 pKa = 4.07HH80 pKa = 6.62GIAFYY85 pKa = 10.45RR86 pKa = 11.84YY87 pKa = 10.1NINTNNWQTLQYY99 pKa = 9.11NPSWATEE106 pKa = 3.96GSDD109 pKa = 4.77DD110 pKa = 4.63GSSLVWAGGDD120 pKa = 3.48YY121 pKa = 10.66LYY123 pKa = 11.2SLQGEE128 pKa = 4.44HH129 pKa = 7.07AEE131 pKa = 4.7SDD133 pKa = 3.87PADD136 pKa = 3.64DD137 pKa = 3.71RR138 pKa = 11.84AFARR142 pKa = 11.84FDD144 pKa = 3.13ITDD147 pKa = 3.74NSWTDD152 pKa = 3.25LDD154 pKa = 5.57DD155 pKa = 4.01IPEE158 pKa = 4.3EE159 pKa = 4.47GGVGDD164 pKa = 4.43GGSLAWAGGNYY175 pKa = 9.37SNKK178 pKa = 9.46VYY180 pKa = 10.9ALGGGFVGRR189 pKa = 11.84GEE191 pKa = 4.27TPGDD195 pKa = 3.15NFYY198 pKa = 11.4VYY200 pKa = 9.17DD201 pKa = 3.89TSVDD205 pKa = 2.81NWTQLQSLPYY215 pKa = 10.89GMTGQNGPRR224 pKa = 11.84LGYY227 pKa = 10.8ADD229 pKa = 3.67GSLYY233 pKa = 10.4CWRR236 pKa = 11.84GYY238 pKa = 11.12YY239 pKa = 9.86DD240 pKa = 3.97ASNNPDD246 pKa = 2.84VLMAYY251 pKa = 7.54EE252 pKa = 4.53LPAVGVSVSISPSEE266 pKa = 3.88NNALNGEE273 pKa = 4.34NVTFTVTVTNVDD285 pKa = 3.98DD286 pKa = 3.74EE287 pKa = 4.48HH288 pKa = 8.42HH289 pKa = 6.6NFVLEE294 pKa = 4.27NSDD297 pKa = 3.69SLDD300 pKa = 3.21WQMSLSEE307 pKa = 5.35DD308 pKa = 3.91NLSLDD313 pKa = 3.97PDD315 pKa = 3.41NSEE318 pKa = 4.04NVVLEE323 pKa = 4.03VAIPPAVSGGTEE335 pKa = 3.93DD336 pKa = 3.8TVTVTATAQDD346 pKa = 3.49NAEE349 pKa = 4.13ISSSAGCTAGAQVDD363 pKa = 3.45VDD365 pKa = 4.1FNVWITPFSQTGSRR379 pKa = 11.84EE380 pKa = 3.96DD381 pKa = 3.35NLDD384 pKa = 3.54YY385 pKa = 11.0VVHH388 pKa = 6.27VKK390 pKa = 10.85NSGNVVDD397 pKa = 5.59NYY399 pKa = 11.7DD400 pKa = 5.98LIITDD405 pKa = 3.56NAGWADD411 pKa = 3.78NVSLAGSGYY420 pKa = 10.66SDD422 pKa = 4.52ADD424 pKa = 3.86NIFPSADD431 pKa = 3.16AGICEE436 pKa = 4.69YY437 pKa = 11.14YY438 pKa = 10.34PNTTKK443 pKa = 10.99GGDD446 pKa = 3.3NYY448 pKa = 11.02NVYY451 pKa = 10.41VGYY454 pKa = 10.52EE455 pKa = 3.91NNPYY459 pKa = 10.29KK460 pKa = 10.38PNCTGEE466 pKa = 3.98DD467 pKa = 3.15TRR469 pKa = 11.84ALFKK473 pKa = 10.67FDD475 pKa = 4.8LSGIPDD481 pKa = 4.13GATITGATLNAHH493 pKa = 6.75GDD495 pKa = 4.09FSPSTGFPNYY505 pKa = 9.6NDD507 pKa = 4.89RR508 pKa = 11.84IMLTDD513 pKa = 3.65VQPLGSDD520 pKa = 2.6SWGEE524 pKa = 3.81NTVNWNNKK532 pKa = 8.55PSFGSTLDD540 pKa = 3.49SVLIDD545 pKa = 4.66DD546 pKa = 4.67NEE548 pKa = 4.06YY549 pKa = 10.34WSWNVKK555 pKa = 10.11SAVQNEE561 pKa = 4.09VSGDD565 pKa = 3.75GVVSLGLLSEE575 pKa = 4.36NAVSNQKK582 pKa = 10.13NVTMWFHH589 pKa = 6.28TKK591 pKa = 10.32DD592 pKa = 3.69GSPEE596 pKa = 3.93NPDD599 pKa = 3.14SVKK602 pKa = 10.47PYY604 pKa = 11.02LEE606 pKa = 4.03VLYY609 pKa = 10.15EE610 pKa = 4.18VYY612 pKa = 10.74DD613 pKa = 3.91VVSLKK618 pKa = 10.84NISPGEE624 pKa = 4.06NEE626 pKa = 3.84NVALNVTIPNSAASGDD642 pKa = 3.77HH643 pKa = 4.73TVKK646 pKa = 10.85VGATSYY652 pKa = 11.78GDD654 pKa = 3.8DD655 pKa = 3.95SLSKK659 pKa = 10.46EE660 pKa = 4.22NSCIASVGSPTGVGVSISPSEE681 pKa = 4.58DD682 pKa = 3.18NAPPGYY688 pKa = 9.89GLNYY692 pKa = 9.84EE693 pKa = 4.33VTVTNTGSISDD704 pKa = 4.19NYY706 pKa = 10.62DD707 pKa = 3.13LSVVDD712 pKa = 4.1NAVPSWNPTLDD723 pKa = 3.14NSLFEE728 pKa = 4.75DD729 pKa = 4.07VPPGEE734 pKa = 4.19NRR736 pKa = 11.84NTVLRR741 pKa = 11.84VTVPEE746 pKa = 3.91NANLGVEE753 pKa = 4.28DD754 pKa = 4.82NIIVTASSQEE764 pKa = 4.02NVEE767 pKa = 3.93MDD769 pKa = 4.53DD770 pKa = 3.76NASCIALSAALFRR783 pKa = 11.84SVKK786 pKa = 10.11ISVSPSYY793 pKa = 10.62KK794 pKa = 10.11AGSSGSSLDD803 pKa = 3.49YY804 pKa = 10.67SVKK807 pKa = 10.26VKK809 pKa = 10.72NRR811 pKa = 11.84GNIPDD816 pKa = 3.88NYY818 pKa = 9.85EE819 pKa = 4.04LEE821 pKa = 4.65ASDD824 pKa = 5.82DD825 pKa = 4.21VFPSWDD831 pKa = 3.45PTVVDD836 pKa = 4.58NIFLNVVPEE845 pKa = 4.17EE846 pKa = 4.16EE847 pKa = 4.38RR848 pKa = 11.84VTTLNVVIPEE858 pKa = 3.96NDD860 pKa = 3.22NLRR863 pKa = 11.84GIEE866 pKa = 4.08DD867 pKa = 4.47NITTTVTSLEE877 pKa = 4.11NDD879 pKa = 3.78SVHH882 pKa = 7.54DD883 pKa = 4.05SAVCTAQAMDD893 pKa = 5.02SISMQLDD900 pKa = 4.07LVADD904 pKa = 4.13WNLVGLPVVSDD915 pKa = 3.49STTKK919 pKa = 10.58EE920 pKa = 3.69NLFEE924 pKa = 4.3GTGVDD929 pKa = 3.75PLSVV933 pKa = 3.14
Molecular weight: 100.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A133VKJ0|A0A133VKJ0_9EURY Uncharacterized protein OS=candidate division MSBL1 archaeon SCGC-AAA382K21 OX=1698283 GN=AKJ54_00950 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 10.04SDD4 pKa = 3.61KK5 pKa = 11.11NKK7 pKa = 10.4GPSITSGKK15 pKa = 8.72TSFRR19 pKa = 11.84SLTGPIHH26 pKa = 7.01SIRR29 pKa = 11.84NLGQLGKK36 pKa = 10.45VRR38 pKa = 11.84TDD40 pKa = 2.82RR41 pKa = 11.84ATLSKK46 pKa = 10.55RR47 pKa = 11.84IFQEE51 pKa = 3.98EE52 pKa = 4.56VVPRR56 pKa = 11.84RR57 pKa = 11.84VGGLAVKK64 pKa = 10.67AFGDD68 pKa = 3.79LRR70 pKa = 11.84LDD72 pKa = 3.82SPQDD76 pKa = 3.35TKK78 pKa = 11.17KK79 pKa = 10.74YY80 pKa = 8.15SQEE83 pKa = 3.49RR84 pKa = 11.84GKK86 pKa = 10.83RR87 pKa = 11.84IIEE90 pKa = 4.38DD91 pKa = 3.48NSRR94 pKa = 11.84LDD96 pKa = 3.44PVFRR100 pKa = 11.84PVGG103 pKa = 3.36
MM1 pKa = 7.67KK2 pKa = 10.04SDD4 pKa = 3.61KK5 pKa = 11.11NKK7 pKa = 10.4GPSITSGKK15 pKa = 8.72TSFRR19 pKa = 11.84SLTGPIHH26 pKa = 7.01SIRR29 pKa = 11.84NLGQLGKK36 pKa = 10.45VRR38 pKa = 11.84TDD40 pKa = 2.82RR41 pKa = 11.84ATLSKK46 pKa = 10.55RR47 pKa = 11.84IFQEE51 pKa = 3.98EE52 pKa = 4.56VVPRR56 pKa = 11.84RR57 pKa = 11.84VGGLAVKK64 pKa = 10.67AFGDD68 pKa = 3.79LRR70 pKa = 11.84LDD72 pKa = 3.82SPQDD76 pKa = 3.35TKK78 pKa = 11.17KK79 pKa = 10.74YY80 pKa = 8.15SQEE83 pKa = 3.49RR84 pKa = 11.84GKK86 pKa = 10.83RR87 pKa = 11.84IIEE90 pKa = 4.38DD91 pKa = 3.48NSRR94 pKa = 11.84LDD96 pKa = 3.44PVFRR100 pKa = 11.84PVGG103 pKa = 3.36
Molecular weight: 11.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
61194 |
53 |
1467 |
280.7 |
31.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.852 ± 0.155 | 0.833 ± 0.045 |
6.102 ± 0.148 | 9.754 ± 0.273 |
3.798 ± 0.125 | 7.517 ± 0.153 |
1.646 ± 0.066 | 7.002 ± 0.165 |
7.377 ± 0.278 | 9.117 ± 0.207 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.062 ± 0.08 | 4.492 ± 0.188 |
3.852 ± 0.098 | 2.229 ± 0.07 |
5.396 ± 0.169 | 7.12 ± 0.208 |
4.804 ± 0.15 | 6.912 ± 0.184 |
1.159 ± 0.067 | 2.976 ± 0.121 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
