
Apis mellifera associated microvirus 61
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.2
Get precalculated fractions of proteins
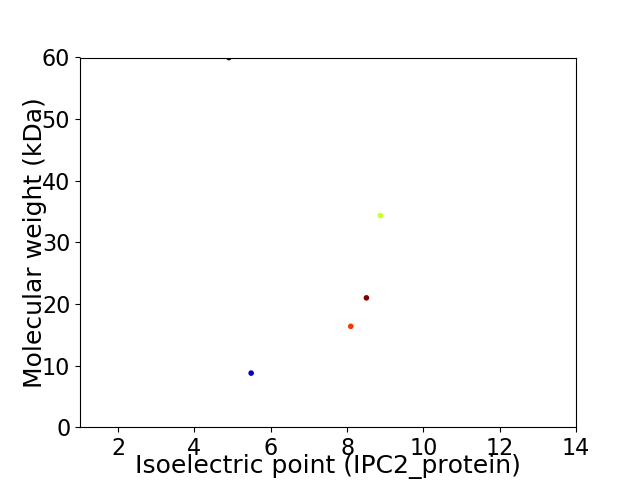
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q8U5U3|A0A3Q8U5U3_9VIRU Replication initiator protein OS=Apis mellifera associated microvirus 61 OX=2494793 PE=4 SV=1
MM1 pKa = 7.18YY2 pKa = 9.62GQSNMHH8 pKa = 5.41VQGPQINQFNNIAPEE23 pKa = 3.94PVPRR27 pKa = 11.84SMFKK31 pKa = 10.32RR32 pKa = 11.84DD33 pKa = 3.3FTRR36 pKa = 11.84RR37 pKa = 11.84STFDD41 pKa = 3.48PDD43 pKa = 3.49YY44 pKa = 10.65LVPIFLDD51 pKa = 4.35EE52 pKa = 4.36ILPGDD57 pKa = 4.0EE58 pKa = 5.06VNLKK62 pKa = 9.21MNAFVRR68 pKa = 11.84MSTPIKK74 pKa = 10.17PLMDD78 pKa = 3.79NLYY81 pKa = 11.01LEE83 pKa = 4.77TFWFFVPNRR92 pKa = 11.84LVWDD96 pKa = 3.44NWEE99 pKa = 4.25KK100 pKa = 10.89FQGQQTNPGDD110 pKa = 4.02SVDD113 pKa = 3.67YY114 pKa = 8.84TIPVLDD120 pKa = 4.26DD121 pKa = 3.37TVAPLSTTGFPVEE134 pKa = 4.48SIYY137 pKa = 10.8DD138 pKa = 3.81YY139 pKa = 11.19FGLPVLVTGIEE150 pKa = 4.64GISCLFNRR158 pKa = 11.84AYY160 pKa = 10.79DD161 pKa = 4.46LIWNEE166 pKa = 3.65WFRR169 pKa = 11.84DD170 pKa = 3.58EE171 pKa = 5.2NLQNSLTVPTGDD183 pKa = 5.25GPDD186 pKa = 3.84DD187 pKa = 3.91PANYY191 pKa = 9.14TLRR194 pKa = 11.84KK195 pKa = 8.2RR196 pKa = 11.84NKK198 pKa = 8.71RR199 pKa = 11.84KK200 pKa = 10.43DD201 pKa = 3.62YY202 pKa = 7.86FTSMLPWPQKK212 pKa = 10.17GDD214 pKa = 3.16AVMIPLAGSAPVISNGQVPKK234 pKa = 10.78LQATSSGNFGNLANSNTIGLTLSSPSGSIGNEE266 pKa = 3.48DD267 pKa = 3.41LRR269 pKa = 11.84FYY271 pKa = 7.67TTNTGLEE278 pKa = 3.97ADD280 pKa = 4.43LDD282 pKa = 4.23AVSALSINVLRR293 pKa = 11.84QSALYY298 pKa = 9.65QQILEE303 pKa = 4.13LDD305 pKa = 3.37ARR307 pKa = 11.84GGTRR311 pKa = 11.84YY312 pKa = 9.89VEE314 pKa = 4.27SVYY317 pKa = 11.12SRR319 pKa = 11.84FGVILPDD326 pKa = 3.83FRR328 pKa = 11.84VQRR331 pKa = 11.84PEE333 pKa = 4.11LLGQGSTRR341 pKa = 11.84INIYY345 pKa = 10.26AVPQTAPSTDD355 pKa = 4.16DD356 pKa = 3.61SPQGNLAAFGTAQISGNHH374 pKa = 5.09GCRR377 pKa = 11.84YY378 pKa = 9.82AATEE382 pKa = 3.79HH383 pKa = 6.22GMLIGLANVRR393 pKa = 11.84ADD395 pKa = 3.35LTYY398 pKa = 10.67QQGVPKK404 pKa = 10.37KK405 pKa = 9.02FLRR408 pKa = 11.84STRR411 pKa = 11.84LDD413 pKa = 3.58FYY415 pKa = 11.56EE416 pKa = 4.36PLQNGLGEE424 pKa = 3.88QSVLNKK430 pKa = 9.93EE431 pKa = 4.98LYY433 pKa = 10.3AQGTSADD440 pKa = 3.96EE441 pKa = 4.41DD442 pKa = 3.85VAGYY446 pKa = 7.16QEE448 pKa = 5.0RR449 pKa = 11.84FAEE452 pKa = 4.15YY453 pKa = 9.56RR454 pKa = 11.84WFNSEE459 pKa = 3.61VTGKK463 pKa = 9.66FRR465 pKa = 11.84SVAPTTLDD473 pKa = 3.04VWHH476 pKa = 7.24LSQEE480 pKa = 4.39FGSLPLLNSSFITEE494 pKa = 4.68DD495 pKa = 3.27VPMDD499 pKa = 3.46RR500 pKa = 11.84VLATGSTEE508 pKa = 3.88PAFLYY513 pKa = 9.92DD514 pKa = 2.93AHH516 pKa = 7.44YY517 pKa = 10.54SYY519 pKa = 9.76THH521 pKa = 5.65VRR523 pKa = 11.84PMPIRR528 pKa = 11.84SNPGIRR534 pKa = 11.84RR535 pKa = 11.84LL536 pKa = 3.55
MM1 pKa = 7.18YY2 pKa = 9.62GQSNMHH8 pKa = 5.41VQGPQINQFNNIAPEE23 pKa = 3.94PVPRR27 pKa = 11.84SMFKK31 pKa = 10.32RR32 pKa = 11.84DD33 pKa = 3.3FTRR36 pKa = 11.84RR37 pKa = 11.84STFDD41 pKa = 3.48PDD43 pKa = 3.49YY44 pKa = 10.65LVPIFLDD51 pKa = 4.35EE52 pKa = 4.36ILPGDD57 pKa = 4.0EE58 pKa = 5.06VNLKK62 pKa = 9.21MNAFVRR68 pKa = 11.84MSTPIKK74 pKa = 10.17PLMDD78 pKa = 3.79NLYY81 pKa = 11.01LEE83 pKa = 4.77TFWFFVPNRR92 pKa = 11.84LVWDD96 pKa = 3.44NWEE99 pKa = 4.25KK100 pKa = 10.89FQGQQTNPGDD110 pKa = 4.02SVDD113 pKa = 3.67YY114 pKa = 8.84TIPVLDD120 pKa = 4.26DD121 pKa = 3.37TVAPLSTTGFPVEE134 pKa = 4.48SIYY137 pKa = 10.8DD138 pKa = 3.81YY139 pKa = 11.19FGLPVLVTGIEE150 pKa = 4.64GISCLFNRR158 pKa = 11.84AYY160 pKa = 10.79DD161 pKa = 4.46LIWNEE166 pKa = 3.65WFRR169 pKa = 11.84DD170 pKa = 3.58EE171 pKa = 5.2NLQNSLTVPTGDD183 pKa = 5.25GPDD186 pKa = 3.84DD187 pKa = 3.91PANYY191 pKa = 9.14TLRR194 pKa = 11.84KK195 pKa = 8.2RR196 pKa = 11.84NKK198 pKa = 8.71RR199 pKa = 11.84KK200 pKa = 10.43DD201 pKa = 3.62YY202 pKa = 7.86FTSMLPWPQKK212 pKa = 10.17GDD214 pKa = 3.16AVMIPLAGSAPVISNGQVPKK234 pKa = 10.78LQATSSGNFGNLANSNTIGLTLSSPSGSIGNEE266 pKa = 3.48DD267 pKa = 3.41LRR269 pKa = 11.84FYY271 pKa = 7.67TTNTGLEE278 pKa = 3.97ADD280 pKa = 4.43LDD282 pKa = 4.23AVSALSINVLRR293 pKa = 11.84QSALYY298 pKa = 9.65QQILEE303 pKa = 4.13LDD305 pKa = 3.37ARR307 pKa = 11.84GGTRR311 pKa = 11.84YY312 pKa = 9.89VEE314 pKa = 4.27SVYY317 pKa = 11.12SRR319 pKa = 11.84FGVILPDD326 pKa = 3.83FRR328 pKa = 11.84VQRR331 pKa = 11.84PEE333 pKa = 4.11LLGQGSTRR341 pKa = 11.84INIYY345 pKa = 10.26AVPQTAPSTDD355 pKa = 4.16DD356 pKa = 3.61SPQGNLAAFGTAQISGNHH374 pKa = 5.09GCRR377 pKa = 11.84YY378 pKa = 9.82AATEE382 pKa = 3.79HH383 pKa = 6.22GMLIGLANVRR393 pKa = 11.84ADD395 pKa = 3.35LTYY398 pKa = 10.67QQGVPKK404 pKa = 10.37KK405 pKa = 9.02FLRR408 pKa = 11.84STRR411 pKa = 11.84LDD413 pKa = 3.58FYY415 pKa = 11.56EE416 pKa = 4.36PLQNGLGEE424 pKa = 3.88QSVLNKK430 pKa = 9.93EE431 pKa = 4.98LYY433 pKa = 10.3AQGTSADD440 pKa = 3.96EE441 pKa = 4.41DD442 pKa = 3.85VAGYY446 pKa = 7.16QEE448 pKa = 5.0RR449 pKa = 11.84FAEE452 pKa = 4.15YY453 pKa = 9.56RR454 pKa = 11.84WFNSEE459 pKa = 3.61VTGKK463 pKa = 9.66FRR465 pKa = 11.84SVAPTTLDD473 pKa = 3.04VWHH476 pKa = 7.24LSQEE480 pKa = 4.39FGSLPLLNSSFITEE494 pKa = 4.68DD495 pKa = 3.27VPMDD499 pKa = 3.46RR500 pKa = 11.84VLATGSTEE508 pKa = 3.88PAFLYY513 pKa = 9.92DD514 pKa = 2.93AHH516 pKa = 7.44YY517 pKa = 10.54SYY519 pKa = 9.76THH521 pKa = 5.65VRR523 pKa = 11.84PMPIRR528 pKa = 11.84SNPGIRR534 pKa = 11.84RR535 pKa = 11.84LL536 pKa = 3.55
Molecular weight: 59.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UU75|A0A3S8UU75_9VIRU DNA pilot protein OS=Apis mellifera associated microvirus 61 OX=2494793 PE=4 SV=1
MM1 pKa = 7.23CTRR4 pKa = 11.84EE5 pKa = 3.44ITAYY9 pKa = 10.1QNPSGGRR16 pKa = 11.84PIFGWEE22 pKa = 4.09GKK24 pKa = 10.47KK25 pKa = 10.47NGLPEE30 pKa = 4.65LKK32 pKa = 10.01LPCGKK37 pKa = 10.38CPEE40 pKa = 4.36CCKK43 pKa = 10.63DD44 pKa = 4.42YY45 pKa = 9.19YY46 pKa = 10.63TSWATRR52 pKa = 11.84GSRR55 pKa = 11.84EE56 pKa = 3.78LALWDD61 pKa = 3.33STVFITLTYY70 pKa = 10.22SDD72 pKa = 4.06EE73 pKa = 4.41HH74 pKa = 7.04LPPDD78 pKa = 3.38NSLNKK83 pKa = 9.86KK84 pKa = 9.86DD85 pKa = 3.52VQDD88 pKa = 4.49FIKK91 pKa = 10.4RR92 pKa = 11.84VKK94 pKa = 10.46KK95 pKa = 10.22RR96 pKa = 11.84FKK98 pKa = 9.49STKK101 pKa = 9.07EE102 pKa = 3.54NPIRR106 pKa = 11.84QIYY109 pKa = 8.75CGEE112 pKa = 4.02YY113 pKa = 10.16GEE115 pKa = 4.43KK116 pKa = 9.45TKK118 pKa = 10.66RR119 pKa = 11.84PHH121 pKa = 4.64YY122 pKa = 10.11HH123 pKa = 7.34AILFNCDD130 pKa = 3.08FADD133 pKa = 3.94KK134 pKa = 9.97KK135 pKa = 8.98PHH137 pKa = 5.53YY138 pKa = 8.51MSPQGHH144 pKa = 5.03QVYY147 pKa = 8.62TSQTLNNLWKK157 pKa = 10.88NGFAEE162 pKa = 4.81FGYY165 pKa = 10.3AQPGSIAYY173 pKa = 9.3LFKK176 pKa = 11.16YY177 pKa = 9.53VLKK180 pKa = 10.64KK181 pKa = 10.33KK182 pKa = 9.66SRR184 pKa = 11.84KK185 pKa = 8.82EE186 pKa = 3.68KK187 pKa = 9.35EE188 pKa = 3.65QPLIIEE194 pKa = 4.51RR195 pKa = 11.84DD196 pKa = 3.51GVTYY200 pKa = 10.25EE201 pKa = 4.05VAHH204 pKa = 6.02EE205 pKa = 4.42FIEE208 pKa = 4.36ASRR211 pKa = 11.84NPGIGAHH218 pKa = 5.6MRR220 pKa = 11.84NSDD223 pKa = 4.31SIKK226 pKa = 10.48KK227 pKa = 10.51GYY229 pKa = 8.67LTVNGVKK236 pKa = 10.27KK237 pKa = 10.52KK238 pKa = 10.08LSKK241 pKa = 11.08YY242 pKa = 8.01YY243 pKa = 10.8LEE245 pKa = 4.31WLRR248 pKa = 11.84KK249 pKa = 9.28NDD251 pKa = 4.07PDD253 pKa = 3.95TFDD256 pKa = 4.85NISNMKK262 pKa = 9.84FDD264 pKa = 5.24FMCKK268 pKa = 9.74LPKK271 pKa = 9.9EE272 pKa = 4.03SRR274 pKa = 11.84LRR276 pKa = 11.84KK277 pKa = 7.57EE278 pKa = 4.43QKK280 pKa = 10.03EE281 pKa = 4.07KK282 pKa = 10.79AQKK285 pKa = 10.87KK286 pKa = 8.08LTDD289 pKa = 3.65TKK291 pKa = 11.23KK292 pKa = 10.9KK293 pKa = 10.52LL294 pKa = 3.39
MM1 pKa = 7.23CTRR4 pKa = 11.84EE5 pKa = 3.44ITAYY9 pKa = 10.1QNPSGGRR16 pKa = 11.84PIFGWEE22 pKa = 4.09GKK24 pKa = 10.47KK25 pKa = 10.47NGLPEE30 pKa = 4.65LKK32 pKa = 10.01LPCGKK37 pKa = 10.38CPEE40 pKa = 4.36CCKK43 pKa = 10.63DD44 pKa = 4.42YY45 pKa = 9.19YY46 pKa = 10.63TSWATRR52 pKa = 11.84GSRR55 pKa = 11.84EE56 pKa = 3.78LALWDD61 pKa = 3.33STVFITLTYY70 pKa = 10.22SDD72 pKa = 4.06EE73 pKa = 4.41HH74 pKa = 7.04LPPDD78 pKa = 3.38NSLNKK83 pKa = 9.86KK84 pKa = 9.86DD85 pKa = 3.52VQDD88 pKa = 4.49FIKK91 pKa = 10.4RR92 pKa = 11.84VKK94 pKa = 10.46KK95 pKa = 10.22RR96 pKa = 11.84FKK98 pKa = 9.49STKK101 pKa = 9.07EE102 pKa = 3.54NPIRR106 pKa = 11.84QIYY109 pKa = 8.75CGEE112 pKa = 4.02YY113 pKa = 10.16GEE115 pKa = 4.43KK116 pKa = 9.45TKK118 pKa = 10.66RR119 pKa = 11.84PHH121 pKa = 4.64YY122 pKa = 10.11HH123 pKa = 7.34AILFNCDD130 pKa = 3.08FADD133 pKa = 3.94KK134 pKa = 9.97KK135 pKa = 8.98PHH137 pKa = 5.53YY138 pKa = 8.51MSPQGHH144 pKa = 5.03QVYY147 pKa = 8.62TSQTLNNLWKK157 pKa = 10.88NGFAEE162 pKa = 4.81FGYY165 pKa = 10.3AQPGSIAYY173 pKa = 9.3LFKK176 pKa = 11.16YY177 pKa = 9.53VLKK180 pKa = 10.64KK181 pKa = 10.33KK182 pKa = 9.66SRR184 pKa = 11.84KK185 pKa = 8.82EE186 pKa = 3.68KK187 pKa = 9.35EE188 pKa = 3.65QPLIIEE194 pKa = 4.51RR195 pKa = 11.84DD196 pKa = 3.51GVTYY200 pKa = 10.25EE201 pKa = 4.05VAHH204 pKa = 6.02EE205 pKa = 4.42FIEE208 pKa = 4.36ASRR211 pKa = 11.84NPGIGAHH218 pKa = 5.6MRR220 pKa = 11.84NSDD223 pKa = 4.31SIKK226 pKa = 10.48KK227 pKa = 10.51GYY229 pKa = 8.67LTVNGVKK236 pKa = 10.27KK237 pKa = 10.52KK238 pKa = 10.08LSKK241 pKa = 11.08YY242 pKa = 8.01YY243 pKa = 10.8LEE245 pKa = 4.31WLRR248 pKa = 11.84KK249 pKa = 9.28NDD251 pKa = 4.07PDD253 pKa = 3.95TFDD256 pKa = 4.85NISNMKK262 pKa = 9.84FDD264 pKa = 5.24FMCKK268 pKa = 9.74LPKK271 pKa = 9.9EE272 pKa = 4.03SRR274 pKa = 11.84LRR276 pKa = 11.84KK277 pKa = 7.57EE278 pKa = 4.43QKK280 pKa = 10.03EE281 pKa = 4.07KK282 pKa = 10.79AQKK285 pKa = 10.87KK286 pKa = 8.08LTDD289 pKa = 3.65TKK291 pKa = 11.23KK292 pKa = 10.9KK293 pKa = 10.52LL294 pKa = 3.39
Molecular weight: 34.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1253 |
77 |
536 |
250.6 |
28.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.704 ± 1.39 | 0.878 ± 0.526 |
6.464 ± 0.821 | 5.347 ± 0.506 |
4.469 ± 0.618 | 7.103 ± 0.398 |
1.596 ± 0.299 | 5.108 ± 0.614 |
8.14 ± 2.769 | 7.901 ± 0.856 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.995 ± 0.127 | 5.427 ± 0.453 |
6.145 ± 0.762 | 5.347 ± 0.959 |
4.549 ± 0.764 | 7.023 ± 0.513 |
6.145 ± 0.359 | 4.868 ± 0.861 |
1.117 ± 0.303 | 3.671 ± 0.71 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
