
Acinetobacter phage phiAB6
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Beijerinckvirinae; Friunavirus; Acinetobacter virus phiAB6
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
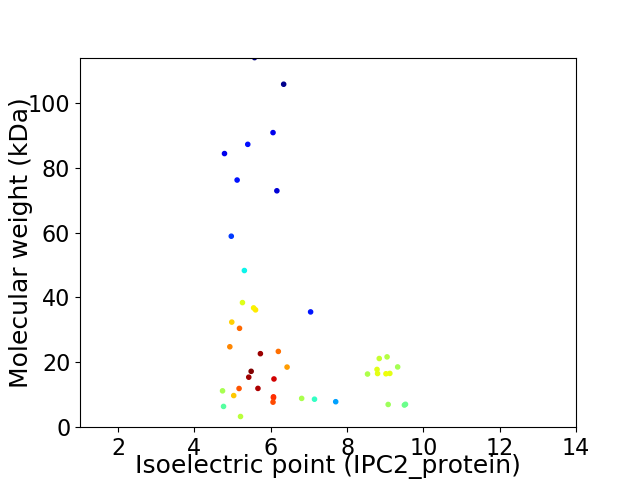
Virtual 2D-PAGE plot for 45 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A159BDB5|A0A159BDB5_9CAUD Tail fiber OS=Acinetobacter phage phiAB6 OX=1698439 GN=phiAB6_gp40 PE=1 SV=1
MM1 pKa = 8.01ILEE4 pKa = 4.25GVYY7 pKa = 10.26PSFLKK12 pKa = 10.85GVSQQAPQEE21 pKa = 4.16RR22 pKa = 11.84SDD24 pKa = 3.82GQLGAQLNLLSDD36 pKa = 3.92AVTGLRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84GGVKK48 pKa = 10.14FQAKK52 pKa = 8.46LTGIPNSSYY61 pKa = 10.72IRR63 pKa = 11.84LIDD66 pKa = 3.58INGVNYY72 pKa = 10.13IMIVDD77 pKa = 4.67TVTGTLKK84 pKa = 10.3IYY86 pKa = 10.89NFDD89 pKa = 3.51GSLLKK94 pKa = 10.64AHH96 pKa = 5.61QTDD99 pKa = 3.84YY100 pKa = 11.69LKK102 pKa = 11.05ASNGKK107 pKa = 9.45ASIRR111 pKa = 11.84STVSRR116 pKa = 11.84NNCFVLNTEE125 pKa = 4.09QVITKK130 pKa = 8.94TPTGGTNPIPNPSTMGYY147 pKa = 9.4ISIRR151 pKa = 11.84SGQFSKK157 pKa = 10.19MYY159 pKa = 10.62SVDD162 pKa = 3.38IKK164 pKa = 10.92SGSYY168 pKa = 8.9TLSFGVGTSGSEE180 pKa = 3.45AWQATPEE187 pKa = 4.08WVATEE192 pKa = 3.73MEE194 pKa = 4.13NRR196 pKa = 11.84IKK198 pKa = 10.89EE199 pKa = 4.23DD200 pKa = 3.05TTLNARR206 pKa = 11.84YY207 pKa = 7.24TVVRR211 pKa = 11.84EE212 pKa = 4.07GSTVALKK219 pKa = 10.76AKK221 pKa = 10.37SAVDD225 pKa = 3.4TNLLVIEE232 pKa = 4.83SGTGSTYY239 pKa = 10.41IQTSNSSRR247 pKa = 11.84VQGKK251 pKa = 8.59QDD253 pKa = 3.83IIANLPNILDD263 pKa = 3.7KK264 pKa = 11.45YY265 pKa = 9.33IIAVGTVGNSAYY277 pKa = 10.01YY278 pKa = 9.98QYY280 pKa = 11.68NATTSTWKK288 pKa = 10.35EE289 pKa = 3.92CGVYY293 pKa = 9.23EE294 pKa = 4.96APYY297 pKa = 10.56KK298 pKa = 9.36FTNEE302 pKa = 3.79PIYY305 pKa = 10.21WYY307 pKa = 10.53FDD309 pKa = 3.43DD310 pKa = 4.54TDD312 pKa = 4.19TIQVKK317 pKa = 10.55SLDD320 pKa = 3.71IQPRR324 pKa = 11.84TAGDD328 pKa = 3.9DD329 pKa = 4.01DD330 pKa = 5.2NNPLPKK336 pKa = 10.25FVDD339 pKa = 4.52FGITGISAYY348 pKa = 8.0QSRR351 pKa = 11.84LVLLSGSYY359 pKa = 11.38VNMSATADD367 pKa = 3.51FNVYY371 pKa = 8.7MRR373 pKa = 11.84TTVEE377 pKa = 4.22EE378 pKa = 4.35LQDD381 pKa = 4.1DD382 pKa = 4.3DD383 pKa = 5.17PIEE386 pKa = 4.31VSSTALSAAQFEE398 pKa = 4.95YY399 pKa = 10.69AVPYY403 pKa = 10.94NKK405 pKa = 10.41DD406 pKa = 3.41LVLLAQNQQAVIPANSTVLTPKK428 pKa = 9.54TAVIYY433 pKa = 9.5PSSKK437 pKa = 11.07ANISMASEE445 pKa = 4.02PQVVSRR451 pKa = 11.84SLYY454 pKa = 8.15YY455 pKa = 9.88TYY457 pKa = 10.89QRR459 pKa = 11.84GTDD462 pKa = 3.74YY463 pKa = 11.16YY464 pKa = 10.96QVGEE468 pKa = 4.81MIPNAYY474 pKa = 10.43SDD476 pKa = 3.65AQYY479 pKa = 10.95YY480 pKa = 8.69AQNLADD486 pKa = 5.62HH487 pKa = 6.88IPLYY491 pKa = 10.03ATGVCTSITGSTTDD505 pKa = 3.0NMAVFSSDD513 pKa = 3.16QKK515 pKa = 11.22EE516 pKa = 4.34LLVHH520 pKa = 5.96QYY522 pKa = 10.11LWAGEE527 pKa = 4.08DD528 pKa = 3.39RR529 pKa = 11.84PLMSFHH535 pKa = 6.66KK536 pKa = 10.23WEE538 pKa = 4.11LPYY541 pKa = 11.02DD542 pKa = 3.81VLHH545 pKa = 5.82VQFLQEE551 pKa = 3.98YY552 pKa = 8.99LVLFMDD558 pKa = 3.9VGDD561 pKa = 4.48DD562 pKa = 3.91LVVGTVNVQLNQLDD576 pKa = 3.96NKK578 pKa = 9.83PIPFLDD584 pKa = 3.25IYY586 pKa = 10.85QYY588 pKa = 11.56VDD590 pKa = 2.84IVDD593 pKa = 4.16GEE595 pKa = 4.56GTLPEE600 pKa = 4.31FLPEE604 pKa = 4.45GEE606 pKa = 4.58LVAAVYY612 pKa = 10.54SSEE615 pKa = 4.19TMRR618 pKa = 11.84HH619 pKa = 5.59AMVQYY624 pKa = 9.82EE625 pKa = 4.19IEE627 pKa = 4.14GTKK630 pKa = 9.98IKK632 pKa = 10.62CQFNGRR638 pKa = 11.84IYY640 pKa = 10.75LGVPYY645 pKa = 10.02EE646 pKa = 4.26SSLTLTPPFVKK657 pKa = 9.94DD658 pKa = 3.39DD659 pKa = 3.6KK660 pKa = 11.07GRR662 pKa = 11.84VVAGSNSTVVDD673 pKa = 3.45LTMTFKK679 pKa = 10.34STGEE683 pKa = 4.05FEE685 pKa = 4.48YY686 pKa = 10.69HH687 pKa = 6.96VSDD690 pKa = 3.56TYY692 pKa = 11.94GDD694 pKa = 3.76VFDD697 pKa = 5.83GEE699 pKa = 4.6TSAQAWSEE707 pKa = 3.85AQLGYY712 pKa = 9.5TRR714 pKa = 11.84VNTVSDD720 pKa = 3.62VKK722 pKa = 10.48FPCGTLLNSTEE733 pKa = 3.9FSIRR737 pKa = 11.84TTGTTEE743 pKa = 3.85LNIISASYY751 pKa = 10.17NIRR754 pKa = 11.84VPNRR758 pKa = 11.84GRR760 pKa = 11.84RR761 pKa = 11.84HH762 pKa = 5.06LL763 pKa = 3.96
MM1 pKa = 8.01ILEE4 pKa = 4.25GVYY7 pKa = 10.26PSFLKK12 pKa = 10.85GVSQQAPQEE21 pKa = 4.16RR22 pKa = 11.84SDD24 pKa = 3.82GQLGAQLNLLSDD36 pKa = 3.92AVTGLRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84GGVKK48 pKa = 10.14FQAKK52 pKa = 8.46LTGIPNSSYY61 pKa = 10.72IRR63 pKa = 11.84LIDD66 pKa = 3.58INGVNYY72 pKa = 10.13IMIVDD77 pKa = 4.67TVTGTLKK84 pKa = 10.3IYY86 pKa = 10.89NFDD89 pKa = 3.51GSLLKK94 pKa = 10.64AHH96 pKa = 5.61QTDD99 pKa = 3.84YY100 pKa = 11.69LKK102 pKa = 11.05ASNGKK107 pKa = 9.45ASIRR111 pKa = 11.84STVSRR116 pKa = 11.84NNCFVLNTEE125 pKa = 4.09QVITKK130 pKa = 8.94TPTGGTNPIPNPSTMGYY147 pKa = 9.4ISIRR151 pKa = 11.84SGQFSKK157 pKa = 10.19MYY159 pKa = 10.62SVDD162 pKa = 3.38IKK164 pKa = 10.92SGSYY168 pKa = 8.9TLSFGVGTSGSEE180 pKa = 3.45AWQATPEE187 pKa = 4.08WVATEE192 pKa = 3.73MEE194 pKa = 4.13NRR196 pKa = 11.84IKK198 pKa = 10.89EE199 pKa = 4.23DD200 pKa = 3.05TTLNARR206 pKa = 11.84YY207 pKa = 7.24TVVRR211 pKa = 11.84EE212 pKa = 4.07GSTVALKK219 pKa = 10.76AKK221 pKa = 10.37SAVDD225 pKa = 3.4TNLLVIEE232 pKa = 4.83SGTGSTYY239 pKa = 10.41IQTSNSSRR247 pKa = 11.84VQGKK251 pKa = 8.59QDD253 pKa = 3.83IIANLPNILDD263 pKa = 3.7KK264 pKa = 11.45YY265 pKa = 9.33IIAVGTVGNSAYY277 pKa = 10.01YY278 pKa = 9.98QYY280 pKa = 11.68NATTSTWKK288 pKa = 10.35EE289 pKa = 3.92CGVYY293 pKa = 9.23EE294 pKa = 4.96APYY297 pKa = 10.56KK298 pKa = 9.36FTNEE302 pKa = 3.79PIYY305 pKa = 10.21WYY307 pKa = 10.53FDD309 pKa = 3.43DD310 pKa = 4.54TDD312 pKa = 4.19TIQVKK317 pKa = 10.55SLDD320 pKa = 3.71IQPRR324 pKa = 11.84TAGDD328 pKa = 3.9DD329 pKa = 4.01DD330 pKa = 5.2NNPLPKK336 pKa = 10.25FVDD339 pKa = 4.52FGITGISAYY348 pKa = 8.0QSRR351 pKa = 11.84LVLLSGSYY359 pKa = 11.38VNMSATADD367 pKa = 3.51FNVYY371 pKa = 8.7MRR373 pKa = 11.84TTVEE377 pKa = 4.22EE378 pKa = 4.35LQDD381 pKa = 4.1DD382 pKa = 4.3DD383 pKa = 5.17PIEE386 pKa = 4.31VSSTALSAAQFEE398 pKa = 4.95YY399 pKa = 10.69AVPYY403 pKa = 10.94NKK405 pKa = 10.41DD406 pKa = 3.41LVLLAQNQQAVIPANSTVLTPKK428 pKa = 9.54TAVIYY433 pKa = 9.5PSSKK437 pKa = 11.07ANISMASEE445 pKa = 4.02PQVVSRR451 pKa = 11.84SLYY454 pKa = 8.15YY455 pKa = 9.88TYY457 pKa = 10.89QRR459 pKa = 11.84GTDD462 pKa = 3.74YY463 pKa = 11.16YY464 pKa = 10.96QVGEE468 pKa = 4.81MIPNAYY474 pKa = 10.43SDD476 pKa = 3.65AQYY479 pKa = 10.95YY480 pKa = 8.69AQNLADD486 pKa = 5.62HH487 pKa = 6.88IPLYY491 pKa = 10.03ATGVCTSITGSTTDD505 pKa = 3.0NMAVFSSDD513 pKa = 3.16QKK515 pKa = 11.22EE516 pKa = 4.34LLVHH520 pKa = 5.96QYY522 pKa = 10.11LWAGEE527 pKa = 4.08DD528 pKa = 3.39RR529 pKa = 11.84PLMSFHH535 pKa = 6.66KK536 pKa = 10.23WEE538 pKa = 4.11LPYY541 pKa = 11.02DD542 pKa = 3.81VLHH545 pKa = 5.82VQFLQEE551 pKa = 3.98YY552 pKa = 8.99LVLFMDD558 pKa = 3.9VGDD561 pKa = 4.48DD562 pKa = 3.91LVVGTVNVQLNQLDD576 pKa = 3.96NKK578 pKa = 9.83PIPFLDD584 pKa = 3.25IYY586 pKa = 10.85QYY588 pKa = 11.56VDD590 pKa = 2.84IVDD593 pKa = 4.16GEE595 pKa = 4.56GTLPEE600 pKa = 4.31FLPEE604 pKa = 4.45GEE606 pKa = 4.58LVAAVYY612 pKa = 10.54SSEE615 pKa = 4.19TMRR618 pKa = 11.84HH619 pKa = 5.59AMVQYY624 pKa = 9.82EE625 pKa = 4.19IEE627 pKa = 4.14GTKK630 pKa = 9.98IKK632 pKa = 10.62CQFNGRR638 pKa = 11.84IYY640 pKa = 10.75LGVPYY645 pKa = 10.02EE646 pKa = 4.26SSLTLTPPFVKK657 pKa = 9.94DD658 pKa = 3.39DD659 pKa = 3.6KK660 pKa = 11.07GRR662 pKa = 11.84VVAGSNSTVVDD673 pKa = 3.45LTMTFKK679 pKa = 10.34STGEE683 pKa = 4.05FEE685 pKa = 4.48YY686 pKa = 10.69HH687 pKa = 6.96VSDD690 pKa = 3.56TYY692 pKa = 11.94GDD694 pKa = 3.76VFDD697 pKa = 5.83GEE699 pKa = 4.6TSAQAWSEE707 pKa = 3.85AQLGYY712 pKa = 9.5TRR714 pKa = 11.84VNTVSDD720 pKa = 3.62VKK722 pKa = 10.48FPCGTLLNSTEE733 pKa = 3.9FSIRR737 pKa = 11.84TTGTTEE743 pKa = 3.85LNIISASYY751 pKa = 10.17NIRR754 pKa = 11.84VPNRR758 pKa = 11.84GRR760 pKa = 11.84RR761 pKa = 11.84HH762 pKa = 5.06LL763 pKa = 3.96
Molecular weight: 84.34 kDa
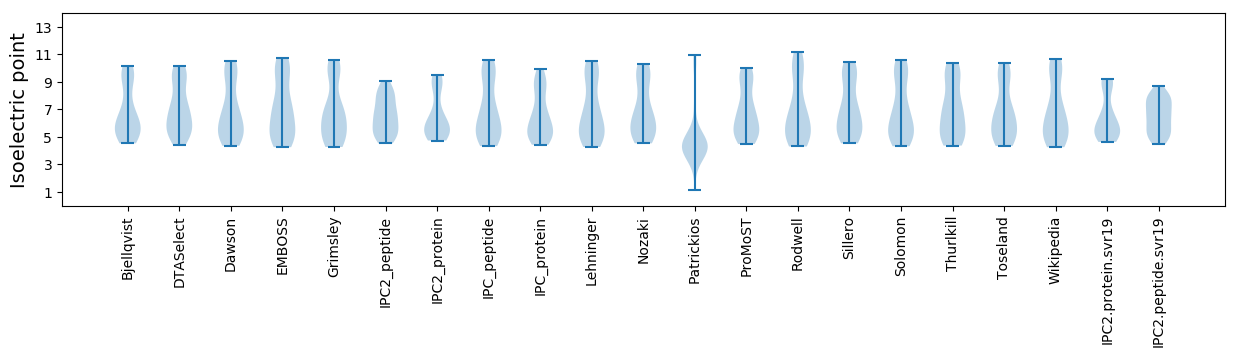
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A159BD85|A0A159BD85_9CAUD Uncharacterized protein OS=Acinetobacter phage phiAB6 OX=1698439 GN=phiAB6_gp9 PE=4 SV=1
MM1 pKa = 7.54SIKK4 pKa = 10.26GYY6 pKa = 9.62SVIQAYY12 pKa = 10.18VDD14 pKa = 3.45EE15 pKa = 4.83LQEE18 pKa = 3.94LTNYY22 pKa = 7.08EE23 pKa = 3.75QRR25 pKa = 11.84KK26 pKa = 8.56RR27 pKa = 11.84EE28 pKa = 3.72QRR30 pKa = 11.84EE31 pKa = 3.52RR32 pKa = 11.84KK33 pKa = 10.27NIVKK37 pKa = 9.48EE38 pKa = 4.0RR39 pKa = 11.84KK40 pKa = 8.94LDD42 pKa = 3.56RR43 pKa = 11.84RR44 pKa = 11.84SKK46 pKa = 11.0RR47 pKa = 11.84EE48 pKa = 3.63AKK50 pKa = 10.09RR51 pKa = 11.84NAWVV55 pKa = 2.99
MM1 pKa = 7.54SIKK4 pKa = 10.26GYY6 pKa = 9.62SVIQAYY12 pKa = 10.18VDD14 pKa = 3.45EE15 pKa = 4.83LQEE18 pKa = 3.94LTNYY22 pKa = 7.08EE23 pKa = 3.75QRR25 pKa = 11.84KK26 pKa = 8.56RR27 pKa = 11.84EE28 pKa = 3.72QRR30 pKa = 11.84EE31 pKa = 3.52RR32 pKa = 11.84KK33 pKa = 10.27NIVKK37 pKa = 9.48EE38 pKa = 4.0RR39 pKa = 11.84KK40 pKa = 8.94LDD42 pKa = 3.56RR43 pKa = 11.84RR44 pKa = 11.84SKK46 pKa = 11.0RR47 pKa = 11.84EE48 pKa = 3.63AKK50 pKa = 10.09RR51 pKa = 11.84NAWVV55 pKa = 2.99
Molecular weight: 6.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12132 |
27 |
1032 |
269.6 |
30.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.333 ± 0.375 | 1.063 ± 0.173 |
6.264 ± 0.2 | 6.009 ± 0.262 |
3.429 ± 0.165 | 7.402 ± 0.319 |
2.11 ± 0.198 | 5.638 ± 0.247 |
6.644 ± 0.328 | 8.259 ± 0.304 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.687 ± 0.182 | 5.045 ± 0.283 |
3.503 ± 0.176 | 4.723 ± 0.286 |
4.822 ± 0.271 | 5.786 ± 0.268 |
6.067 ± 0.291 | 6.668 ± 0.251 |
1.261 ± 0.097 | 4.286 ± 0.236 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
