
Lone star tick rhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Lostrhavirus; Lonestar lostrhavirus
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
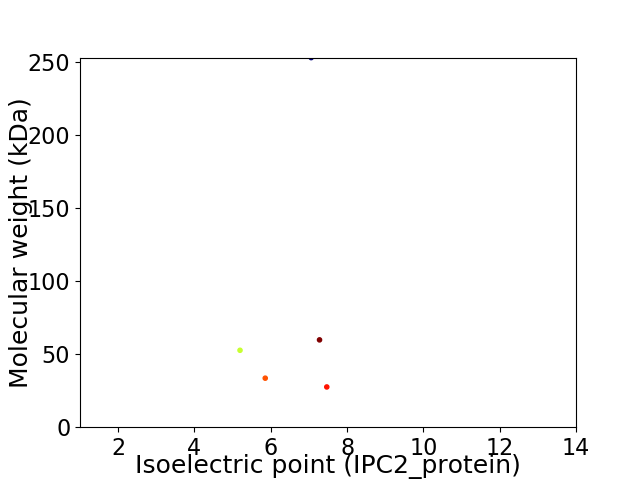
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
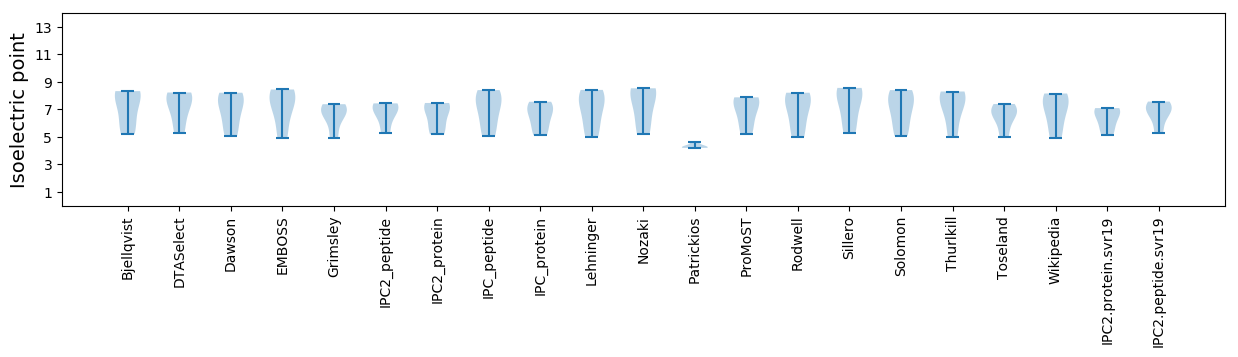
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1N7TF62|A0A1N7TF62_9RHAB Phosphoprotein OS=Lone star tick rhabdovirus OX=1756186 PE=4 SV=1
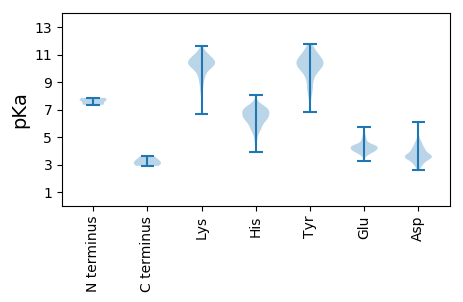
MM1 pKa = 7.34EE2 pKa = 5.48LWPVNVEE9 pKa = 4.38KK10 pKa = 10.92INFRR14 pKa = 11.84PIVTPVTTEE23 pKa = 3.71TAPTYY28 pKa = 9.82PSVWFDD34 pKa = 2.69AHH36 pKa = 6.99ANQKK40 pKa = 10.09PGITARR46 pKa = 11.84VGEE49 pKa = 4.49ANLAYY54 pKa = 10.48HH55 pKa = 6.34NMIIMLEE62 pKa = 4.02QQQWTEE68 pKa = 4.09LTVVSYY74 pKa = 8.17MVAVIRR80 pKa = 11.84EE81 pKa = 4.06NPNIFSTVPTAAWAANGRR99 pKa = 11.84PVARR103 pKa = 11.84ANQRR107 pKa = 11.84VSVLDD112 pKa = 3.36ILTVTNGAAWAAVQQARR129 pKa = 11.84ASTRR133 pKa = 11.84EE134 pKa = 3.89SRR136 pKa = 11.84LSLFWATLLLYY147 pKa = 10.41RR148 pKa = 11.84KK149 pKa = 7.29TLARR153 pKa = 11.84EE154 pKa = 3.65VDD156 pKa = 3.21RR157 pKa = 11.84EE158 pKa = 4.15YY159 pKa = 11.46GNKK162 pKa = 9.4IARR165 pKa = 11.84QLNDD169 pKa = 3.21FFKK172 pKa = 11.12GGVLQMDD179 pKa = 3.45VDD181 pKa = 4.07DD182 pKa = 4.59TEE184 pKa = 4.56EE185 pKa = 5.02DD186 pKa = 3.75GLKK189 pKa = 10.53LGKK192 pKa = 10.14DD193 pKa = 3.43KK194 pKa = 11.61LKK196 pKa = 10.92DD197 pKa = 3.59GYY199 pKa = 10.65LKK201 pKa = 10.76LVAAIDD207 pKa = 3.63MFFFKK212 pKa = 11.01YY213 pKa = 9.53QDD215 pKa = 3.44HH216 pKa = 6.98EE217 pKa = 4.53YY218 pKa = 11.43SMVRR222 pKa = 11.84MATLPSRR229 pKa = 11.84FKK231 pKa = 10.54GCAAITSAGQVAKK244 pKa = 10.51ILGIEE249 pKa = 4.09PGEE252 pKa = 4.34LGLFVYY258 pKa = 8.35TEE260 pKa = 4.1PVAVEE265 pKa = 4.19LANSNRR271 pKa = 11.84PNQGMEE277 pKa = 3.94DD278 pKa = 3.7LYY280 pKa = 11.23GYY282 pKa = 10.19FPYY285 pKa = 10.75QMDD288 pKa = 2.86MGMTQRR294 pKa = 11.84SAYY297 pKa = 9.95SISSNPTLYY306 pKa = 10.8LWLHH310 pKa = 6.47TIGCTVGFQRR320 pKa = 11.84SFQARR325 pKa = 11.84YY326 pKa = 8.92PEE328 pKa = 4.12AAGDD332 pKa = 3.53ISGTMISAAAVGLAFSKK349 pKa = 10.02GLKK352 pKa = 8.53MRR354 pKa = 11.84RR355 pKa = 11.84QFGTTEE361 pKa = 3.62EE362 pKa = 4.04AANALRR368 pKa = 11.84ARR370 pKa = 11.84NQAIDD375 pKa = 3.57QAEE378 pKa = 3.84AAAAEE383 pKa = 4.56VGEE386 pKa = 4.65EE387 pKa = 4.45APPPPPPAQEE397 pKa = 4.02GVEE400 pKa = 4.06AVADD404 pKa = 4.25GGDD407 pKa = 3.88GEE409 pKa = 4.85GDD411 pKa = 3.31AAALDD416 pKa = 4.13IPPPPAEE423 pKa = 4.42EE424 pKa = 4.2NPLAAMSNTILDD436 pKa = 3.75NLIEE440 pKa = 4.38NEE442 pKa = 4.22GLTKK446 pKa = 10.91DD447 pKa = 3.2EE448 pKa = 4.11MKK450 pKa = 10.45EE451 pKa = 3.73FRR453 pKa = 11.84RR454 pKa = 11.84RR455 pKa = 11.84LILGRR460 pKa = 11.84TLRR463 pKa = 11.84EE464 pKa = 3.93TTVGRR469 pKa = 11.84RR470 pKa = 11.84LANIYY475 pKa = 10.77NPGAA479 pKa = 3.62
MM1 pKa = 7.34EE2 pKa = 5.48LWPVNVEE9 pKa = 4.38KK10 pKa = 10.92INFRR14 pKa = 11.84PIVTPVTTEE23 pKa = 3.71TAPTYY28 pKa = 9.82PSVWFDD34 pKa = 2.69AHH36 pKa = 6.99ANQKK40 pKa = 10.09PGITARR46 pKa = 11.84VGEE49 pKa = 4.49ANLAYY54 pKa = 10.48HH55 pKa = 6.34NMIIMLEE62 pKa = 4.02QQQWTEE68 pKa = 4.09LTVVSYY74 pKa = 8.17MVAVIRR80 pKa = 11.84EE81 pKa = 4.06NPNIFSTVPTAAWAANGRR99 pKa = 11.84PVARR103 pKa = 11.84ANQRR107 pKa = 11.84VSVLDD112 pKa = 3.36ILTVTNGAAWAAVQQARR129 pKa = 11.84ASTRR133 pKa = 11.84EE134 pKa = 3.89SRR136 pKa = 11.84LSLFWATLLLYY147 pKa = 10.41RR148 pKa = 11.84KK149 pKa = 7.29TLARR153 pKa = 11.84EE154 pKa = 3.65VDD156 pKa = 3.21RR157 pKa = 11.84EE158 pKa = 4.15YY159 pKa = 11.46GNKK162 pKa = 9.4IARR165 pKa = 11.84QLNDD169 pKa = 3.21FFKK172 pKa = 11.12GGVLQMDD179 pKa = 3.45VDD181 pKa = 4.07DD182 pKa = 4.59TEE184 pKa = 4.56EE185 pKa = 5.02DD186 pKa = 3.75GLKK189 pKa = 10.53LGKK192 pKa = 10.14DD193 pKa = 3.43KK194 pKa = 11.61LKK196 pKa = 10.92DD197 pKa = 3.59GYY199 pKa = 10.65LKK201 pKa = 10.76LVAAIDD207 pKa = 3.63MFFFKK212 pKa = 11.01YY213 pKa = 9.53QDD215 pKa = 3.44HH216 pKa = 6.98EE217 pKa = 4.53YY218 pKa = 11.43SMVRR222 pKa = 11.84MATLPSRR229 pKa = 11.84FKK231 pKa = 10.54GCAAITSAGQVAKK244 pKa = 10.51ILGIEE249 pKa = 4.09PGEE252 pKa = 4.34LGLFVYY258 pKa = 8.35TEE260 pKa = 4.1PVAVEE265 pKa = 4.19LANSNRR271 pKa = 11.84PNQGMEE277 pKa = 3.94DD278 pKa = 3.7LYY280 pKa = 11.23GYY282 pKa = 10.19FPYY285 pKa = 10.75QMDD288 pKa = 2.86MGMTQRR294 pKa = 11.84SAYY297 pKa = 9.95SISSNPTLYY306 pKa = 10.8LWLHH310 pKa = 6.47TIGCTVGFQRR320 pKa = 11.84SFQARR325 pKa = 11.84YY326 pKa = 8.92PEE328 pKa = 4.12AAGDD332 pKa = 3.53ISGTMISAAAVGLAFSKK349 pKa = 10.02GLKK352 pKa = 8.53MRR354 pKa = 11.84RR355 pKa = 11.84QFGTTEE361 pKa = 3.62EE362 pKa = 4.04AANALRR368 pKa = 11.84ARR370 pKa = 11.84NQAIDD375 pKa = 3.57QAEE378 pKa = 3.84AAAAEE383 pKa = 4.56VGEE386 pKa = 4.65EE387 pKa = 4.45APPPPPPAQEE397 pKa = 4.02GVEE400 pKa = 4.06AVADD404 pKa = 4.25GGDD407 pKa = 3.88GEE409 pKa = 4.85GDD411 pKa = 3.31AAALDD416 pKa = 4.13IPPPPAEE423 pKa = 4.42EE424 pKa = 4.2NPLAAMSNTILDD436 pKa = 3.75NLIEE440 pKa = 4.38NEE442 pKa = 4.22GLTKK446 pKa = 10.91DD447 pKa = 3.2EE448 pKa = 4.11MKK450 pKa = 10.45EE451 pKa = 3.73FRR453 pKa = 11.84RR454 pKa = 11.84RR455 pKa = 11.84LILGRR460 pKa = 11.84TLRR463 pKa = 11.84EE464 pKa = 3.93TTVGRR469 pKa = 11.84RR470 pKa = 11.84LANIYY475 pKa = 10.77NPGAA479 pKa = 3.62
Molecular weight: 52.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1N7TF64|A0A1N7TF64_9RHAB Glycoprotein OS=Lone star tick rhabdovirus OX=1756186 PE=3 SV=1
MM1 pKa = 7.7KK2 pKa = 9.22KK3 pKa = 6.64TTNLIHH9 pKa = 7.17IEE11 pKa = 4.06LDD13 pKa = 3.28SLRR16 pKa = 11.84VVSVIRR22 pKa = 11.84SIMLRR27 pKa = 11.84PIRR30 pKa = 11.84DD31 pKa = 2.91AWKK34 pKa = 10.12KK35 pKa = 9.64IKK37 pKa = 10.77KK38 pKa = 9.53NDD40 pKa = 3.53EE41 pKa = 4.02GTTAPAPSAPSTSQTLTMEE60 pKa = 4.32HH61 pKa = 7.46PYY63 pKa = 11.11LEE65 pKa = 4.59GVTVNVPPPLNPGRR79 pKa = 11.84VITDD83 pKa = 4.03LNLEE87 pKa = 4.6LDD89 pKa = 4.13FKK91 pKa = 11.29MEE93 pKa = 4.02VVTNKK98 pKa = 9.64VEE100 pKa = 3.95TDD102 pKa = 3.62LARR105 pKa = 11.84ISAPLINLQWDD116 pKa = 3.74YY117 pKa = 11.75KK118 pKa = 11.28GDD120 pKa = 3.68LRR122 pKa = 11.84YY123 pKa = 10.63KK124 pKa = 9.37NIWMAFAMVSAFGLEE139 pKa = 4.44GGDD142 pKa = 3.66PQPYY146 pKa = 8.15GCLYY150 pKa = 10.72KK151 pKa = 9.98MEE153 pKa = 4.34ICRR156 pKa = 11.84GIKK159 pKa = 9.83IKK161 pKa = 10.97VEE163 pKa = 3.84SSRR166 pKa = 11.84LFEE169 pKa = 4.22TPRR172 pKa = 11.84EE173 pKa = 4.35FKK175 pKa = 10.01WNQYY179 pKa = 7.11VHH181 pKa = 6.85WLADD185 pKa = 4.59GIHH188 pKa = 6.03SEE190 pKa = 3.92WTFRR194 pKa = 11.84GQAIRR199 pKa = 11.84TCVNYY204 pKa = 10.12KK205 pKa = 10.21AQEE208 pKa = 3.98AKK210 pKa = 10.47KK211 pKa = 10.8GLEE214 pKa = 3.76DD215 pKa = 3.4WLNSMGVPARR225 pKa = 11.84RR226 pKa = 11.84DD227 pKa = 3.67EE228 pKa = 4.22NKK230 pKa = 10.42HH231 pKa = 6.35LILIIDD237 pKa = 3.74QEE239 pKa = 4.25QQ240 pKa = 2.9
MM1 pKa = 7.7KK2 pKa = 9.22KK3 pKa = 6.64TTNLIHH9 pKa = 7.17IEE11 pKa = 4.06LDD13 pKa = 3.28SLRR16 pKa = 11.84VVSVIRR22 pKa = 11.84SIMLRR27 pKa = 11.84PIRR30 pKa = 11.84DD31 pKa = 2.91AWKK34 pKa = 10.12KK35 pKa = 9.64IKK37 pKa = 10.77KK38 pKa = 9.53NDD40 pKa = 3.53EE41 pKa = 4.02GTTAPAPSAPSTSQTLTMEE60 pKa = 4.32HH61 pKa = 7.46PYY63 pKa = 11.11LEE65 pKa = 4.59GVTVNVPPPLNPGRR79 pKa = 11.84VITDD83 pKa = 4.03LNLEE87 pKa = 4.6LDD89 pKa = 4.13FKK91 pKa = 11.29MEE93 pKa = 4.02VVTNKK98 pKa = 9.64VEE100 pKa = 3.95TDD102 pKa = 3.62LARR105 pKa = 11.84ISAPLINLQWDD116 pKa = 3.74YY117 pKa = 11.75KK118 pKa = 11.28GDD120 pKa = 3.68LRR122 pKa = 11.84YY123 pKa = 10.63KK124 pKa = 9.37NIWMAFAMVSAFGLEE139 pKa = 4.44GGDD142 pKa = 3.66PQPYY146 pKa = 8.15GCLYY150 pKa = 10.72KK151 pKa = 9.98MEE153 pKa = 4.34ICRR156 pKa = 11.84GIKK159 pKa = 9.83IKK161 pKa = 10.97VEE163 pKa = 3.84SSRR166 pKa = 11.84LFEE169 pKa = 4.22TPRR172 pKa = 11.84EE173 pKa = 4.35FKK175 pKa = 10.01WNQYY179 pKa = 7.11VHH181 pKa = 6.85WLADD185 pKa = 4.59GIHH188 pKa = 6.03SEE190 pKa = 3.92WTFRR194 pKa = 11.84GQAIRR199 pKa = 11.84TCVNYY204 pKa = 10.12KK205 pKa = 10.21AQEE208 pKa = 3.98AKK210 pKa = 10.47KK211 pKa = 10.8GLEE214 pKa = 3.76DD215 pKa = 3.4WLNSMGVPARR225 pKa = 11.84RR226 pKa = 11.84DD227 pKa = 3.67EE228 pKa = 4.22NKK230 pKa = 10.42HH231 pKa = 6.35LILIIDD237 pKa = 3.74QEE239 pKa = 4.25QQ240 pKa = 2.9
Molecular weight: 27.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3742 |
240 |
2195 |
748.4 |
85.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.864 ± 1.406 | 1.523 ± 0.327 |
5.425 ± 0.475 | 6.36 ± 0.246 |
3.661 ± 0.392 | 5.559 ± 0.309 |
2.539 ± 0.399 | 7.376 ± 0.562 |
6.601 ± 0.629 | 9.594 ± 0.797 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.352 ± 0.292 | 4.703 ± 0.309 |
5.158 ± 0.636 | 3.447 ± 0.233 |
5.104 ± 0.352 | 7.402 ± 0.663 |
7.429 ± 0.527 | 5.131 ± 0.407 |
1.871 ± 0.259 | 3.902 ± 0.477 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
