
Herbaspirillum sp. YR522
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Oxalobacteraceae; Herbaspirillum; unclassified Herbaspirillum
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
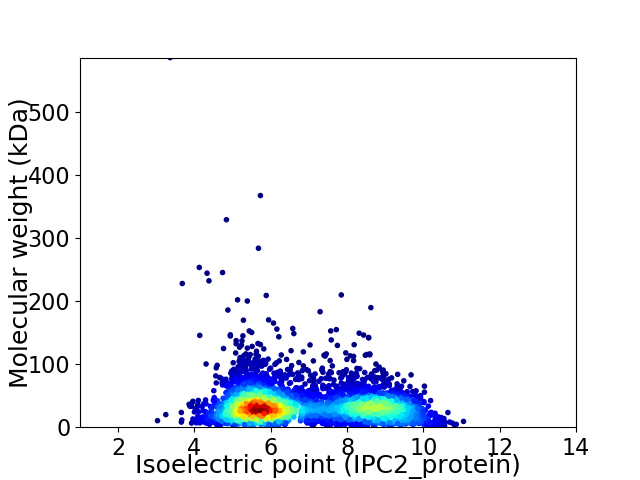
Virtual 2D-PAGE plot for 4612 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J2W9X1|J2W9X1_9BURK Putative pterin-4-alpha-carbinolamine dehydratase OS=Herbaspirillum sp. YR522 OX=1144342 GN=PMI40_00523 PE=3 SV=1
MM1 pKa = 7.15ATAPSPQANPGDD13 pKa = 4.23KK14 pKa = 9.18VTGPVRR20 pKa = 11.84VVMPDD25 pKa = 3.02QGGSFQLNAPRR36 pKa = 11.84GAVQKK41 pKa = 10.17VQVVDD46 pKa = 3.47VDD48 pKa = 4.73LVLHH52 pKa = 6.11MADD55 pKa = 3.19GTKK58 pKa = 10.33VVLAGGAMEE67 pKa = 5.75AMDD70 pKa = 5.38DD71 pKa = 3.49KK72 pKa = 11.73SKK74 pKa = 11.46VKK76 pKa = 10.54FSDD79 pKa = 3.62GSADD83 pKa = 3.29TGRR86 pKa = 11.84LLDD89 pKa = 3.9TVGKK93 pKa = 10.39IPLAPNDD100 pKa = 3.3QSRR103 pKa = 11.84VLNSDD108 pKa = 3.37PVAAQDD114 pKa = 3.48RR115 pKa = 11.84TGTTEE120 pKa = 3.64QYY122 pKa = 11.41GPGQGQGADD131 pKa = 3.14RR132 pKa = 11.84NAAPVASDD140 pKa = 3.9TLSQLAKK147 pKa = 10.5VIADD151 pKa = 3.51NSHH154 pKa = 6.38SVTGDD159 pKa = 3.0ATSQLAKK166 pKa = 10.72VIADD170 pKa = 3.41NSKK173 pKa = 10.77SVTNGAQTLSIPTQPTAGDD192 pKa = 3.71ASPVGGQVATNPNQNSSDD210 pKa = 4.04SEE212 pKa = 3.96PTRR215 pKa = 11.84PVDD218 pKa = 3.45VQVPPEE224 pKa = 4.38RR225 pKa = 11.84PPTTPSTVVTGPGSPTLSVNLVNLTTITQVGDD257 pKa = 3.31VLYY260 pKa = 10.74GSGGTPQSASDD271 pKa = 3.82GSNAVQFARR280 pKa = 11.84QVITAGDD287 pKa = 4.05DD288 pKa = 3.14VHH290 pKa = 6.89TIYY293 pKa = 11.18ASGNNSNTFIKK304 pKa = 10.37VFNVKK309 pKa = 10.13VGGDD313 pKa = 3.57VTLLTLTVMGVPSDD327 pKa = 3.69VTLLNGTNMGGGVYY341 pKa = 10.02QITIAPGQTEE351 pKa = 4.26FNLQFQYY358 pKa = 9.52NTQLANASSPVHH370 pKa = 5.71SQFDD374 pKa = 3.82LSFVTTMATSSGVLTLSDD392 pKa = 3.55TRR394 pKa = 11.84HH395 pKa = 5.07VAIKK399 pKa = 10.39DD400 pKa = 3.58ATSSADD406 pKa = 3.56LTYY409 pKa = 10.78LDD411 pKa = 5.36PSTGDD416 pKa = 3.16NVLIFPAQGVPHH428 pKa = 6.7EE429 pKa = 4.23VHH431 pKa = 6.59AGNGGVTIYY440 pKa = 10.95GSNANDD446 pKa = 3.94LLYY449 pKa = 10.99GGTGNDD455 pKa = 3.28IIYY458 pKa = 10.27GGVGSSSFFQGGAGADD474 pKa = 3.47QLVGSSTGNNTASYY488 pKa = 10.3AGSTQGVTIDD498 pKa = 4.14LNTGVGTGGDD508 pKa = 3.53AQGDD512 pKa = 3.86QLTNIQNLVGSANNDD527 pKa = 3.21TFVASGQVNRR537 pKa = 11.84FDD539 pKa = 4.19GGTGGSDD546 pKa = 3.15TVSYY550 pKa = 10.48AHH552 pKa = 6.6SASGVTVDD560 pKa = 4.46LVNNVGSGGDD570 pKa = 3.57AAGDD574 pKa = 3.6TYY576 pKa = 11.72VSIEE580 pKa = 4.01NVIGSTSNDD589 pKa = 2.96TFIASSAANSFTGNGGTDD607 pKa = 3.22TVSYY611 pKa = 10.55ALSTAGVTVNLKK623 pKa = 10.88SNLGSGGFAAGDD635 pKa = 3.86TYY637 pKa = 11.69NGIQNFVGSAYY648 pKa = 10.59DD649 pKa = 3.44DD650 pKa = 3.81TFVGNAAVNSFDD662 pKa = 4.34GGSGGNDD669 pKa = 2.89TVSYY673 pKa = 10.26AGSTAAVTVNLVTGRR688 pKa = 11.84GSNGDD693 pKa = 3.68AAGDD697 pKa = 3.64TYY699 pKa = 11.29IRR701 pKa = 11.84IQNVIGGDD709 pKa = 3.7GNDD712 pKa = 3.55TFVVGLDD719 pKa = 3.33SKK721 pKa = 11.31NFNGGGGTLNTVDD734 pKa = 3.91YY735 pKa = 9.84SGSSSAITANMLTNTVTRR753 pKa = 11.84TFEE756 pKa = 4.04PVNALTNIQKK766 pKa = 9.92IVGSSYY772 pKa = 10.85SDD774 pKa = 3.34TFIGGAGADD783 pKa = 3.39IFDD786 pKa = 4.65GGLAGSDD793 pKa = 3.37TVDD796 pKa = 3.52YY797 pKa = 10.94SAGAAVTVDD806 pKa = 4.31LFTGTGVGGNAQGDD820 pKa = 4.36TLVHH824 pKa = 5.63IQNVIGSSAGGNTFYY839 pKa = 11.43ASADD843 pKa = 3.41ANGFNGNSTSGNTVNYY859 pKa = 9.8SHH861 pKa = 6.99ATGAVIVDD869 pKa = 3.96LTNTRR874 pKa = 11.84DD875 pKa = 3.2NTLAQNFATGDD886 pKa = 3.58TFTNIQNVVGSVYY899 pKa = 10.95DD900 pKa = 4.0DD901 pKa = 5.29LIVATGQVNRR911 pKa = 11.84IDD913 pKa = 4.19GGLGNNTVSYY923 pKa = 9.29EE924 pKa = 4.01QALTAVKK931 pKa = 10.53VDD933 pKa = 3.91LATNQGSLGDD943 pKa = 4.08AAGDD947 pKa = 3.53TYY949 pKa = 11.81LNIQNAIGGAGNDD962 pKa = 3.26ILVGNASVNKK972 pKa = 9.69FDD974 pKa = 5.44GGLGSNTVSYY984 pKa = 10.07EE985 pKa = 3.81HH986 pKa = 6.6ATAAVKK992 pKa = 10.83VNLASGLGLSGDD1004 pKa = 3.96AAGDD1008 pKa = 3.64TYY1010 pKa = 12.0ANIQNAIGGAGDD1022 pKa = 4.5DD1023 pKa = 4.42IIVASAVANAIDD1035 pKa = 3.91GGGGSGNTVSYY1046 pKa = 9.8EE1047 pKa = 3.87NSSAGVKK1054 pKa = 10.04VDD1056 pKa = 4.08LFLGQGFNNDD1066 pKa = 3.62AAGDD1070 pKa = 3.93TYY1072 pKa = 11.69VRR1074 pKa = 11.84IQNAQGGNGDD1084 pKa = 3.67DD1085 pKa = 3.77TFVGNADD1092 pKa = 3.46VNNFEE1097 pKa = 5.0GGNGSNTVSYY1107 pKa = 9.22EE1108 pKa = 3.91HH1109 pKa = 7.01SSAAVKK1115 pKa = 10.47INLATGSGSLGDD1127 pKa = 3.84AAGDD1131 pKa = 3.17RR1132 pKa = 11.84YY1133 pKa = 11.56YY1134 pKa = 11.07NIQNAIGGLGNDD1146 pKa = 4.39TIVGSAATNAIDD1158 pKa = 4.53GGAGVNTVSYY1168 pKa = 9.78EE1169 pKa = 3.99NSTGAVKK1176 pKa = 10.85VNLATGQGLLGDD1188 pKa = 4.84ALNDD1192 pKa = 3.58TYY1194 pKa = 11.79VRR1196 pKa = 11.84IQNVTGGAGNDD1207 pKa = 3.43ILVGSADD1214 pKa = 3.51VNVIDD1219 pKa = 5.82GGLGNNTVSYY1229 pKa = 9.88EE1230 pKa = 4.15SSTGAVTIDD1239 pKa = 3.73LKK1241 pKa = 10.74QAQGLAGDD1249 pKa = 4.39AAGDD1253 pKa = 3.74TYY1255 pKa = 11.52FNIQNATGGAGDD1267 pKa = 4.22DD1268 pKa = 4.7LIVGSSAVNVIKK1280 pKa = 10.81GGAGVNTVSYY1290 pKa = 8.5ATSTAGVKK1298 pKa = 10.05VDD1300 pKa = 3.9LANNIGTGGDD1310 pKa = 3.67ANLDD1314 pKa = 3.61TYY1316 pKa = 11.69EE1317 pKa = 4.82NIQKK1321 pKa = 10.57VIGSALDD1328 pKa = 3.43DD1329 pKa = 3.74TFVGNGQLNEE1339 pKa = 4.09FWGGTGTNTVSYY1351 pKa = 9.0EE1352 pKa = 3.88ASTGAVTIDD1361 pKa = 3.97LGQQIGTAGDD1371 pKa = 3.48AAGDD1375 pKa = 4.23LYY1377 pKa = 11.31HH1378 pKa = 7.82DD1379 pKa = 4.25IQNATGGAGNDD1390 pKa = 4.21LIIGSAVANRR1400 pKa = 11.84IDD1402 pKa = 4.17GGAGNNTVSYY1412 pKa = 9.58EE1413 pKa = 3.96NSTAAVKK1420 pKa = 10.87VNLVSNQGSLGDD1432 pKa = 3.95AAGDD1436 pKa = 3.58TYY1438 pKa = 11.94VNIQNAIGGAGDD1450 pKa = 5.14DD1451 pKa = 3.39IFVGNGVANRR1461 pKa = 11.84FDD1463 pKa = 3.55GRR1465 pKa = 11.84AGSNTISYY1473 pKa = 8.73EE1474 pKa = 3.93SSGSVTVDD1482 pKa = 3.36LNLGTGTAGDD1492 pKa = 4.18ANGDD1496 pKa = 3.26IYY1498 pKa = 11.65LNIQNFLGGAADD1510 pKa = 3.85DD1511 pKa = 4.13TVVASSVANVIDD1523 pKa = 3.91GGGTSLHH1530 pKa = 6.0NRR1532 pKa = 11.84VSYY1535 pKa = 10.37AASTAAVTVDD1545 pKa = 3.95LNYY1548 pKa = 10.4TDD1550 pKa = 3.85GTGTSGGYY1558 pKa = 9.79AAGDD1562 pKa = 3.68KK1563 pKa = 10.72LSNIQDD1569 pKa = 3.93LTGSIYY1575 pKa = 10.82DD1576 pKa = 3.68DD1577 pKa = 3.67TFVASAAANRR1587 pKa = 11.84FDD1589 pKa = 4.11GGGTSSHH1596 pKa = 5.75NRR1598 pKa = 11.84VSYY1601 pKa = 10.08AASNAGVTVDD1611 pKa = 5.08LNYY1614 pKa = 10.45TDD1616 pKa = 3.85GTGTSGGYY1624 pKa = 9.78AAGDD1628 pKa = 3.58TLVNIQDD1635 pKa = 4.2LTGSAYY1641 pKa = 10.88NDD1643 pKa = 3.55LFVASAASNNFDD1655 pKa = 3.94GGAGSNTVSYY1665 pKa = 10.19AASTAAVTVNLSPSVGSGNGGYY1687 pKa = 10.36AAGDD1691 pKa = 3.63TFTNIQNVIGSAYY1704 pKa = 9.89ADD1706 pKa = 3.7TLIGYY1711 pKa = 7.51ATVGVSSVLTGGAGGDD1727 pKa = 3.7TLIGIAANRR1736 pKa = 11.84AYY1738 pKa = 7.34TTASYY1743 pKa = 10.35AGSSAGVTVDD1753 pKa = 4.62LNYY1756 pKa = 10.68SDD1758 pKa = 4.06GTGTSGGDD1766 pKa = 3.2ATGDD1770 pKa = 2.89RR1771 pKa = 11.84LSFINNLIGSSFNDD1785 pKa = 3.35TFVANAQANSFDD1797 pKa = 4.04GGAGTDD1803 pKa = 3.46TVSYY1807 pKa = 8.65ATSTSAVTVDD1817 pKa = 4.48LSVTDD1822 pKa = 4.24GSGTSGGYY1830 pKa = 9.56AAGDD1834 pKa = 2.94RR1835 pKa = 11.84FVNIEE1840 pKa = 3.98NLIGSAYY1847 pKa = 10.78NDD1849 pKa = 3.7VLKK1852 pKa = 10.91GAAGGNSVIVGGGGADD1868 pKa = 3.61NITGVGTGNYY1878 pKa = 9.62ASYY1881 pKa = 10.17VGSGAAVTIDD1891 pKa = 3.33LRR1893 pKa = 11.84TGVGTGGDD1901 pKa = 3.53AQGDD1905 pKa = 3.86QLNNIYY1911 pKa = 10.73NVIGSSFNDD1920 pKa = 3.38TFIASSQANVFVGGGGVDD1938 pKa = 3.37TVSYY1942 pKa = 10.08IGSTAGVTVDD1952 pKa = 4.69LSNTTGANTSGGYY1965 pKa = 9.82AAGDD1969 pKa = 3.71SFNGISNIVGSNFSDD1984 pKa = 3.23TFYY1987 pKa = 11.51ASAVANFFDD1996 pKa = 5.07GGSGTNTVNYY2006 pKa = 9.75ARR2008 pKa = 11.84STAGVTVDD2016 pKa = 4.4LFHH2019 pKa = 6.57NTGANGYY2026 pKa = 9.55AAGDD2030 pKa = 3.75TYY2032 pKa = 12.01ANIQNATGSAFADD2045 pKa = 3.72LFYY2048 pKa = 11.31ANSSGNAFVGGGGSDD2063 pKa = 3.29TVSYY2067 pKa = 10.82QYY2069 pKa = 10.82ATGSAITASLASGTGSGGDD2088 pKa = 3.51AAGDD2092 pKa = 3.76TYY2094 pKa = 11.63SGIFNLIGSASVNSTLTGDD2113 pKa = 3.88GNGNTLTALGTGTTNVLNAGNAASGQTDD2141 pKa = 4.46IINAQQGGMNTLYY2154 pKa = 10.72AGTGTSTFNVSAGGVNNGIVNASTGTDD2181 pKa = 3.68NLNHH2185 pKa = 7.09AYY2187 pKa = 10.13GAGSAGTTTLHH2198 pKa = 6.81FDD2200 pKa = 3.3NLAATLTLSKK2210 pKa = 10.37FSADD2214 pKa = 3.0ATGISVLDD2222 pKa = 3.76VSSGSNTNMLVRR2234 pKa = 11.84ATDD2237 pKa = 3.57VTSMGVNNVLTIKK2250 pKa = 8.17MTATEE2255 pKa = 4.78SIQIDD2260 pKa = 4.01TTDD2263 pKa = 2.83TAYY2266 pKa = 10.88SYY2268 pKa = 11.15LIFGNGDD2275 pKa = 3.55YY2276 pKa = 11.11VFYY2279 pKa = 10.84AAADD2283 pKa = 3.85TNHH2286 pKa = 5.4VNEE2289 pKa = 4.36VARR2292 pKa = 11.84IHH2294 pKa = 6.14IQLAAA2299 pKa = 3.95
MM1 pKa = 7.15ATAPSPQANPGDD13 pKa = 4.23KK14 pKa = 9.18VTGPVRR20 pKa = 11.84VVMPDD25 pKa = 3.02QGGSFQLNAPRR36 pKa = 11.84GAVQKK41 pKa = 10.17VQVVDD46 pKa = 3.47VDD48 pKa = 4.73LVLHH52 pKa = 6.11MADD55 pKa = 3.19GTKK58 pKa = 10.33VVLAGGAMEE67 pKa = 5.75AMDD70 pKa = 5.38DD71 pKa = 3.49KK72 pKa = 11.73SKK74 pKa = 11.46VKK76 pKa = 10.54FSDD79 pKa = 3.62GSADD83 pKa = 3.29TGRR86 pKa = 11.84LLDD89 pKa = 3.9TVGKK93 pKa = 10.39IPLAPNDD100 pKa = 3.3QSRR103 pKa = 11.84VLNSDD108 pKa = 3.37PVAAQDD114 pKa = 3.48RR115 pKa = 11.84TGTTEE120 pKa = 3.64QYY122 pKa = 11.41GPGQGQGADD131 pKa = 3.14RR132 pKa = 11.84NAAPVASDD140 pKa = 3.9TLSQLAKK147 pKa = 10.5VIADD151 pKa = 3.51NSHH154 pKa = 6.38SVTGDD159 pKa = 3.0ATSQLAKK166 pKa = 10.72VIADD170 pKa = 3.41NSKK173 pKa = 10.77SVTNGAQTLSIPTQPTAGDD192 pKa = 3.71ASPVGGQVATNPNQNSSDD210 pKa = 4.04SEE212 pKa = 3.96PTRR215 pKa = 11.84PVDD218 pKa = 3.45VQVPPEE224 pKa = 4.38RR225 pKa = 11.84PPTTPSTVVTGPGSPTLSVNLVNLTTITQVGDD257 pKa = 3.31VLYY260 pKa = 10.74GSGGTPQSASDD271 pKa = 3.82GSNAVQFARR280 pKa = 11.84QVITAGDD287 pKa = 4.05DD288 pKa = 3.14VHH290 pKa = 6.89TIYY293 pKa = 11.18ASGNNSNTFIKK304 pKa = 10.37VFNVKK309 pKa = 10.13VGGDD313 pKa = 3.57VTLLTLTVMGVPSDD327 pKa = 3.69VTLLNGTNMGGGVYY341 pKa = 10.02QITIAPGQTEE351 pKa = 4.26FNLQFQYY358 pKa = 9.52NTQLANASSPVHH370 pKa = 5.71SQFDD374 pKa = 3.82LSFVTTMATSSGVLTLSDD392 pKa = 3.55TRR394 pKa = 11.84HH395 pKa = 5.07VAIKK399 pKa = 10.39DD400 pKa = 3.58ATSSADD406 pKa = 3.56LTYY409 pKa = 10.78LDD411 pKa = 5.36PSTGDD416 pKa = 3.16NVLIFPAQGVPHH428 pKa = 6.7EE429 pKa = 4.23VHH431 pKa = 6.59AGNGGVTIYY440 pKa = 10.95GSNANDD446 pKa = 3.94LLYY449 pKa = 10.99GGTGNDD455 pKa = 3.28IIYY458 pKa = 10.27GGVGSSSFFQGGAGADD474 pKa = 3.47QLVGSSTGNNTASYY488 pKa = 10.3AGSTQGVTIDD498 pKa = 4.14LNTGVGTGGDD508 pKa = 3.53AQGDD512 pKa = 3.86QLTNIQNLVGSANNDD527 pKa = 3.21TFVASGQVNRR537 pKa = 11.84FDD539 pKa = 4.19GGTGGSDD546 pKa = 3.15TVSYY550 pKa = 10.48AHH552 pKa = 6.6SASGVTVDD560 pKa = 4.46LVNNVGSGGDD570 pKa = 3.57AAGDD574 pKa = 3.6TYY576 pKa = 11.72VSIEE580 pKa = 4.01NVIGSTSNDD589 pKa = 2.96TFIASSAANSFTGNGGTDD607 pKa = 3.22TVSYY611 pKa = 10.55ALSTAGVTVNLKK623 pKa = 10.88SNLGSGGFAAGDD635 pKa = 3.86TYY637 pKa = 11.69NGIQNFVGSAYY648 pKa = 10.59DD649 pKa = 3.44DD650 pKa = 3.81TFVGNAAVNSFDD662 pKa = 4.34GGSGGNDD669 pKa = 2.89TVSYY673 pKa = 10.26AGSTAAVTVNLVTGRR688 pKa = 11.84GSNGDD693 pKa = 3.68AAGDD697 pKa = 3.64TYY699 pKa = 11.29IRR701 pKa = 11.84IQNVIGGDD709 pKa = 3.7GNDD712 pKa = 3.55TFVVGLDD719 pKa = 3.33SKK721 pKa = 11.31NFNGGGGTLNTVDD734 pKa = 3.91YY735 pKa = 9.84SGSSSAITANMLTNTVTRR753 pKa = 11.84TFEE756 pKa = 4.04PVNALTNIQKK766 pKa = 9.92IVGSSYY772 pKa = 10.85SDD774 pKa = 3.34TFIGGAGADD783 pKa = 3.39IFDD786 pKa = 4.65GGLAGSDD793 pKa = 3.37TVDD796 pKa = 3.52YY797 pKa = 10.94SAGAAVTVDD806 pKa = 4.31LFTGTGVGGNAQGDD820 pKa = 4.36TLVHH824 pKa = 5.63IQNVIGSSAGGNTFYY839 pKa = 11.43ASADD843 pKa = 3.41ANGFNGNSTSGNTVNYY859 pKa = 9.8SHH861 pKa = 6.99ATGAVIVDD869 pKa = 3.96LTNTRR874 pKa = 11.84DD875 pKa = 3.2NTLAQNFATGDD886 pKa = 3.58TFTNIQNVVGSVYY899 pKa = 10.95DD900 pKa = 4.0DD901 pKa = 5.29LIVATGQVNRR911 pKa = 11.84IDD913 pKa = 4.19GGLGNNTVSYY923 pKa = 9.29EE924 pKa = 4.01QALTAVKK931 pKa = 10.53VDD933 pKa = 3.91LATNQGSLGDD943 pKa = 4.08AAGDD947 pKa = 3.53TYY949 pKa = 11.81LNIQNAIGGAGNDD962 pKa = 3.26ILVGNASVNKK972 pKa = 9.69FDD974 pKa = 5.44GGLGSNTVSYY984 pKa = 10.07EE985 pKa = 3.81HH986 pKa = 6.6ATAAVKK992 pKa = 10.83VNLASGLGLSGDD1004 pKa = 3.96AAGDD1008 pKa = 3.64TYY1010 pKa = 12.0ANIQNAIGGAGDD1022 pKa = 4.5DD1023 pKa = 4.42IIVASAVANAIDD1035 pKa = 3.91GGGGSGNTVSYY1046 pKa = 9.8EE1047 pKa = 3.87NSSAGVKK1054 pKa = 10.04VDD1056 pKa = 4.08LFLGQGFNNDD1066 pKa = 3.62AAGDD1070 pKa = 3.93TYY1072 pKa = 11.69VRR1074 pKa = 11.84IQNAQGGNGDD1084 pKa = 3.67DD1085 pKa = 3.77TFVGNADD1092 pKa = 3.46VNNFEE1097 pKa = 5.0GGNGSNTVSYY1107 pKa = 9.22EE1108 pKa = 3.91HH1109 pKa = 7.01SSAAVKK1115 pKa = 10.47INLATGSGSLGDD1127 pKa = 3.84AAGDD1131 pKa = 3.17RR1132 pKa = 11.84YY1133 pKa = 11.56YY1134 pKa = 11.07NIQNAIGGLGNDD1146 pKa = 4.39TIVGSAATNAIDD1158 pKa = 4.53GGAGVNTVSYY1168 pKa = 9.78EE1169 pKa = 3.99NSTGAVKK1176 pKa = 10.85VNLATGQGLLGDD1188 pKa = 4.84ALNDD1192 pKa = 3.58TYY1194 pKa = 11.79VRR1196 pKa = 11.84IQNVTGGAGNDD1207 pKa = 3.43ILVGSADD1214 pKa = 3.51VNVIDD1219 pKa = 5.82GGLGNNTVSYY1229 pKa = 9.88EE1230 pKa = 4.15SSTGAVTIDD1239 pKa = 3.73LKK1241 pKa = 10.74QAQGLAGDD1249 pKa = 4.39AAGDD1253 pKa = 3.74TYY1255 pKa = 11.52FNIQNATGGAGDD1267 pKa = 4.22DD1268 pKa = 4.7LIVGSSAVNVIKK1280 pKa = 10.81GGAGVNTVSYY1290 pKa = 8.5ATSTAGVKK1298 pKa = 10.05VDD1300 pKa = 3.9LANNIGTGGDD1310 pKa = 3.67ANLDD1314 pKa = 3.61TYY1316 pKa = 11.69EE1317 pKa = 4.82NIQKK1321 pKa = 10.57VIGSALDD1328 pKa = 3.43DD1329 pKa = 3.74TFVGNGQLNEE1339 pKa = 4.09FWGGTGTNTVSYY1351 pKa = 9.0EE1352 pKa = 3.88ASTGAVTIDD1361 pKa = 3.97LGQQIGTAGDD1371 pKa = 3.48AAGDD1375 pKa = 4.23LYY1377 pKa = 11.31HH1378 pKa = 7.82DD1379 pKa = 4.25IQNATGGAGNDD1390 pKa = 4.21LIIGSAVANRR1400 pKa = 11.84IDD1402 pKa = 4.17GGAGNNTVSYY1412 pKa = 9.58EE1413 pKa = 3.96NSTAAVKK1420 pKa = 10.87VNLVSNQGSLGDD1432 pKa = 3.95AAGDD1436 pKa = 3.58TYY1438 pKa = 11.94VNIQNAIGGAGDD1450 pKa = 5.14DD1451 pKa = 3.39IFVGNGVANRR1461 pKa = 11.84FDD1463 pKa = 3.55GRR1465 pKa = 11.84AGSNTISYY1473 pKa = 8.73EE1474 pKa = 3.93SSGSVTVDD1482 pKa = 3.36LNLGTGTAGDD1492 pKa = 4.18ANGDD1496 pKa = 3.26IYY1498 pKa = 11.65LNIQNFLGGAADD1510 pKa = 3.85DD1511 pKa = 4.13TVVASSVANVIDD1523 pKa = 3.91GGGTSLHH1530 pKa = 6.0NRR1532 pKa = 11.84VSYY1535 pKa = 10.37AASTAAVTVDD1545 pKa = 3.95LNYY1548 pKa = 10.4TDD1550 pKa = 3.85GTGTSGGYY1558 pKa = 9.79AAGDD1562 pKa = 3.68KK1563 pKa = 10.72LSNIQDD1569 pKa = 3.93LTGSIYY1575 pKa = 10.82DD1576 pKa = 3.68DD1577 pKa = 3.67TFVASAAANRR1587 pKa = 11.84FDD1589 pKa = 4.11GGGTSSHH1596 pKa = 5.75NRR1598 pKa = 11.84VSYY1601 pKa = 10.08AASNAGVTVDD1611 pKa = 5.08LNYY1614 pKa = 10.45TDD1616 pKa = 3.85GTGTSGGYY1624 pKa = 9.78AAGDD1628 pKa = 3.58TLVNIQDD1635 pKa = 4.2LTGSAYY1641 pKa = 10.88NDD1643 pKa = 3.55LFVASAASNNFDD1655 pKa = 3.94GGAGSNTVSYY1665 pKa = 10.19AASTAAVTVNLSPSVGSGNGGYY1687 pKa = 10.36AAGDD1691 pKa = 3.63TFTNIQNVIGSAYY1704 pKa = 9.89ADD1706 pKa = 3.7TLIGYY1711 pKa = 7.51ATVGVSSVLTGGAGGDD1727 pKa = 3.7TLIGIAANRR1736 pKa = 11.84AYY1738 pKa = 7.34TTASYY1743 pKa = 10.35AGSSAGVTVDD1753 pKa = 4.62LNYY1756 pKa = 10.68SDD1758 pKa = 4.06GTGTSGGDD1766 pKa = 3.2ATGDD1770 pKa = 2.89RR1771 pKa = 11.84LSFINNLIGSSFNDD1785 pKa = 3.35TFVANAQANSFDD1797 pKa = 4.04GGAGTDD1803 pKa = 3.46TVSYY1807 pKa = 8.65ATSTSAVTVDD1817 pKa = 4.48LSVTDD1822 pKa = 4.24GSGTSGGYY1830 pKa = 9.56AAGDD1834 pKa = 2.94RR1835 pKa = 11.84FVNIEE1840 pKa = 3.98NLIGSAYY1847 pKa = 10.78NDD1849 pKa = 3.7VLKK1852 pKa = 10.91GAAGGNSVIVGGGGADD1868 pKa = 3.61NITGVGTGNYY1878 pKa = 9.62ASYY1881 pKa = 10.17VGSGAAVTIDD1891 pKa = 3.33LRR1893 pKa = 11.84TGVGTGGDD1901 pKa = 3.53AQGDD1905 pKa = 3.86QLNNIYY1911 pKa = 10.73NVIGSSFNDD1920 pKa = 3.38TFIASSQANVFVGGGGVDD1938 pKa = 3.37TVSYY1942 pKa = 10.08IGSTAGVTVDD1952 pKa = 4.69LSNTTGANTSGGYY1965 pKa = 9.82AAGDD1969 pKa = 3.71SFNGISNIVGSNFSDD1984 pKa = 3.23TFYY1987 pKa = 11.51ASAVANFFDD1996 pKa = 5.07GGSGTNTVNYY2006 pKa = 9.75ARR2008 pKa = 11.84STAGVTVDD2016 pKa = 4.4LFHH2019 pKa = 6.57NTGANGYY2026 pKa = 9.55AAGDD2030 pKa = 3.75TYY2032 pKa = 12.01ANIQNATGSAFADD2045 pKa = 3.72LFYY2048 pKa = 11.31ANSSGNAFVGGGGSDD2063 pKa = 3.29TVSYY2067 pKa = 10.82QYY2069 pKa = 10.82ATGSAITASLASGTGSGGDD2088 pKa = 3.51AAGDD2092 pKa = 3.76TYY2094 pKa = 11.63SGIFNLIGSASVNSTLTGDD2113 pKa = 3.88GNGNTLTALGTGTTNVLNAGNAASGQTDD2141 pKa = 4.46IINAQQGGMNTLYY2154 pKa = 10.72AGTGTSTFNVSAGGVNNGIVNASTGTDD2181 pKa = 3.68NLNHH2185 pKa = 7.09AYY2187 pKa = 10.13GAGSAGTTTLHH2198 pKa = 6.81FDD2200 pKa = 3.3NLAATLTLSKK2210 pKa = 10.37FSADD2214 pKa = 3.0ATGISVLDD2222 pKa = 3.76VSSGSNTNMLVRR2234 pKa = 11.84ATDD2237 pKa = 3.57VTSMGVNNVLTIKK2250 pKa = 8.17MTATEE2255 pKa = 4.78SIQIDD2260 pKa = 4.01TTDD2263 pKa = 2.83TAYY2266 pKa = 10.88SYY2268 pKa = 11.15LIFGNGDD2275 pKa = 3.55YY2276 pKa = 11.11VFYY2279 pKa = 10.84AAADD2283 pKa = 3.85TNHH2286 pKa = 5.4VNEE2289 pKa = 4.36VARR2292 pKa = 11.84IHH2294 pKa = 6.14IQLAAA2299 pKa = 3.95
Molecular weight: 227.72 kDa
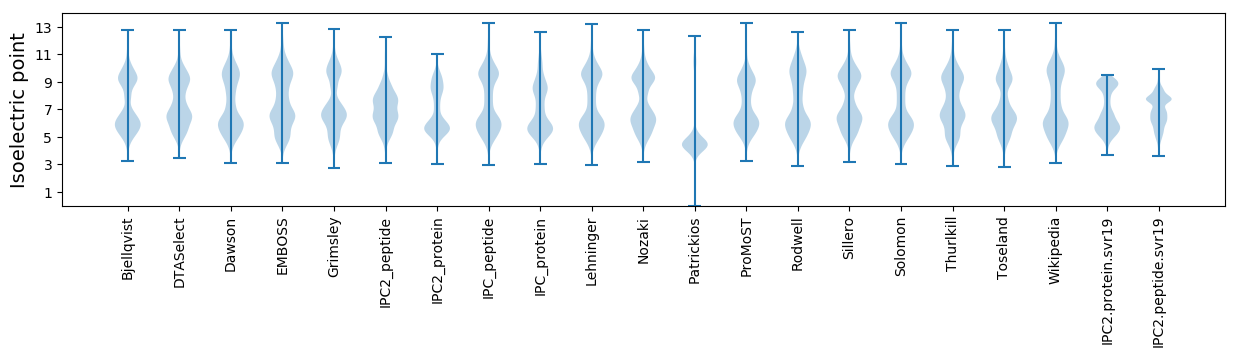
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J2VUT1|J2VUT1_9BURK Putative redox protein regulator of disulfide bond formation OS=Herbaspirillum sp. YR522 OX=1144342 GN=PMI40_00270 PE=4 SV=1
MM1 pKa = 7.65PWLLAVRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.79KK11 pKa = 9.93HH12 pKa = 5.92LRR14 pKa = 11.84LHH16 pKa = 5.01QHH18 pKa = 7.08LLPKK22 pKa = 10.3RR23 pKa = 11.84LPLPQHH29 pKa = 5.78QLPKK33 pKa = 10.47RR34 pKa = 11.84LPLPLLLLKK43 pKa = 10.46RR44 pKa = 11.84LPLLLLLLKK53 pKa = 10.42HH54 pKa = 6.44LLLLPPMQSQLTLHH68 pKa = 6.24LQSNRR73 pKa = 11.84KK74 pKa = 9.31IIFF77 pKa = 3.58
MM1 pKa = 7.65PWLLAVRR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.79KK11 pKa = 9.93HH12 pKa = 5.92LRR14 pKa = 11.84LHH16 pKa = 5.01QHH18 pKa = 7.08LLPKK22 pKa = 10.3RR23 pKa = 11.84LPLPQHH29 pKa = 5.78QLPKK33 pKa = 10.47RR34 pKa = 11.84LPLPLLLLKK43 pKa = 10.46RR44 pKa = 11.84LPLLLLLLKK53 pKa = 10.42HH54 pKa = 6.44LLLLPPMQSQLTLHH68 pKa = 6.24LQSNRR73 pKa = 11.84KK74 pKa = 9.31IIFF77 pKa = 3.58
Molecular weight: 9.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1510888 |
12 |
5836 |
327.6 |
35.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.387 ± 0.048 | 0.838 ± 0.011 |
5.447 ± 0.032 | 4.948 ± 0.037 |
3.51 ± 0.024 | 8.184 ± 0.06 |
2.235 ± 0.018 | 4.922 ± 0.029 |
3.235 ± 0.034 | 10.752 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.551 ± 0.019 | 2.951 ± 0.038 |
4.846 ± 0.034 | 4.496 ± 0.03 |
6.774 ± 0.045 | 5.731 ± 0.03 |
5.059 ± 0.04 | 7.45 ± 0.027 |
1.312 ± 0.015 | 2.372 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
