
Nocardioides silvaticus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
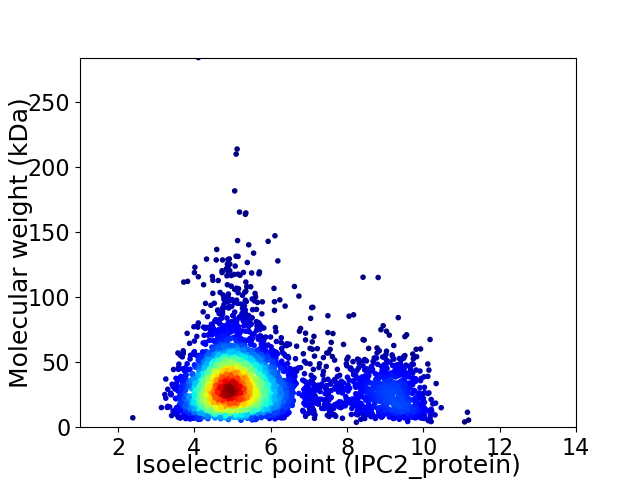
Virtual 2D-PAGE plot for 4220 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A316TQ38|A0A316TQ38_9ACTN DNA-protecting protein DprA OS=Nocardioides silvaticus OX=2201891 GN=dprA PE=3 SV=1
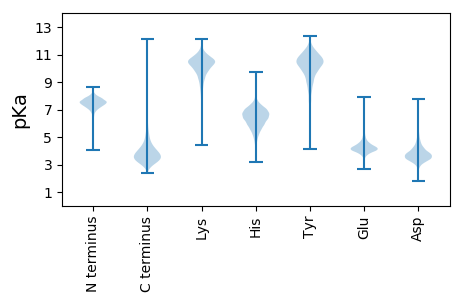
MM1 pKa = 7.21NRR3 pKa = 11.84TGRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84SATVGLWAALTVLAAIVFPSGAQAHH33 pKa = 7.27DD34 pKa = 3.98DD35 pKa = 3.58AGSRR39 pKa = 11.84GLSAGGTHH47 pKa = 6.4TCAIDD52 pKa = 3.42SAGSISCWGYY62 pKa = 11.21DD63 pKa = 4.64DD64 pKa = 5.48YY65 pKa = 11.99GQTTGPDD72 pKa = 3.17GAAGVFKK79 pKa = 10.58AVSAGSSHH87 pKa = 6.19TCAIDD92 pKa = 3.29ASGSISCWGEE102 pKa = 3.32NSYY105 pKa = 11.19GQVSGPDD112 pKa = 3.43AAAGTYY118 pKa = 8.15TAVSAGTSHH127 pKa = 6.47TCAIEE132 pKa = 3.91ATGSISCWGRR142 pKa = 11.84DD143 pKa = 3.36DD144 pKa = 4.43HH145 pKa = 6.87GQVGDD150 pKa = 3.94PDD152 pKa = 3.66AAAGTFKK159 pKa = 10.39TVAADD164 pKa = 3.75NLHH167 pKa = 5.71TCAVGTSGSISCWGDD182 pKa = 3.13DD183 pKa = 3.8SHH185 pKa = 7.04GQTSGPDD192 pKa = 3.34GAAGAFTTVSVRR204 pKa = 11.84YY205 pKa = 8.37FQTCAIDD212 pKa = 3.35VAGAISCWGNNGAGQVDD229 pKa = 4.19APIGAGFKK237 pKa = 9.56TVSLGLFHH245 pKa = 6.89TCAINAADD253 pKa = 6.0SIICWGHH260 pKa = 6.93DD261 pKa = 3.13MFGEE265 pKa = 4.05VDD267 pKa = 4.16GPNSSAGTFKK277 pKa = 10.47TLSLGDD283 pKa = 3.73GHH285 pKa = 6.54TCAIDD290 pKa = 3.45AADD293 pKa = 5.78SISCWGFDD301 pKa = 4.47SGIGWVTGADD311 pKa = 3.44AAPGSFGTTPADD323 pKa = 3.65PEE325 pKa = 4.42CTIGDD330 pKa = 3.88PNSNQRR336 pKa = 11.84QVLRR340 pKa = 11.84GTPGPDD346 pKa = 3.4VMCGGAAADD355 pKa = 4.01SFHH358 pKa = 6.78GAGGEE363 pKa = 4.06DD364 pKa = 3.87IILGGGGNDD373 pKa = 3.38WLNGGTGNDD382 pKa = 3.93TVDD385 pKa = 3.31GGAGQDD391 pKa = 3.51QLRR394 pKa = 11.84GEE396 pKa = 4.63AGADD400 pKa = 3.39ALDD403 pKa = 4.51GGDD406 pKa = 5.18DD407 pKa = 3.71NDD409 pKa = 4.16SVKK412 pKa = 11.04GGVGNDD418 pKa = 3.44TVTGGAGRR426 pKa = 11.84DD427 pKa = 3.68VLDD430 pKa = 3.54EE431 pKa = 4.14TGAAGFGNDD440 pKa = 4.97DD441 pKa = 4.63LDD443 pKa = 5.64GGDD446 pKa = 3.79GNDD449 pKa = 3.62VIRR452 pKa = 11.84GGEE455 pKa = 4.28GVDD458 pKa = 4.41SIAGGDD464 pKa = 3.97GNDD467 pKa = 3.96NLNGGAGSPDD477 pKa = 3.47VCDD480 pKa = 4.63GGPGTDD486 pKa = 2.71IGGYY490 pKa = 9.48GCEE493 pKa = 4.17QKK495 pKa = 11.03VSIPP499 pKa = 3.45
MM1 pKa = 7.21NRR3 pKa = 11.84TGRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84SATVGLWAALTVLAAIVFPSGAQAHH33 pKa = 7.27DD34 pKa = 3.98DD35 pKa = 3.58AGSRR39 pKa = 11.84GLSAGGTHH47 pKa = 6.4TCAIDD52 pKa = 3.42SAGSISCWGYY62 pKa = 11.21DD63 pKa = 4.64DD64 pKa = 5.48YY65 pKa = 11.99GQTTGPDD72 pKa = 3.17GAAGVFKK79 pKa = 10.58AVSAGSSHH87 pKa = 6.19TCAIDD92 pKa = 3.29ASGSISCWGEE102 pKa = 3.32NSYY105 pKa = 11.19GQVSGPDD112 pKa = 3.43AAAGTYY118 pKa = 8.15TAVSAGTSHH127 pKa = 6.47TCAIEE132 pKa = 3.91ATGSISCWGRR142 pKa = 11.84DD143 pKa = 3.36DD144 pKa = 4.43HH145 pKa = 6.87GQVGDD150 pKa = 3.94PDD152 pKa = 3.66AAAGTFKK159 pKa = 10.39TVAADD164 pKa = 3.75NLHH167 pKa = 5.71TCAVGTSGSISCWGDD182 pKa = 3.13DD183 pKa = 3.8SHH185 pKa = 7.04GQTSGPDD192 pKa = 3.34GAAGAFTTVSVRR204 pKa = 11.84YY205 pKa = 8.37FQTCAIDD212 pKa = 3.35VAGAISCWGNNGAGQVDD229 pKa = 4.19APIGAGFKK237 pKa = 9.56TVSLGLFHH245 pKa = 6.89TCAINAADD253 pKa = 6.0SIICWGHH260 pKa = 6.93DD261 pKa = 3.13MFGEE265 pKa = 4.05VDD267 pKa = 4.16GPNSSAGTFKK277 pKa = 10.47TLSLGDD283 pKa = 3.73GHH285 pKa = 6.54TCAIDD290 pKa = 3.45AADD293 pKa = 5.78SISCWGFDD301 pKa = 4.47SGIGWVTGADD311 pKa = 3.44AAPGSFGTTPADD323 pKa = 3.65PEE325 pKa = 4.42CTIGDD330 pKa = 3.88PNSNQRR336 pKa = 11.84QVLRR340 pKa = 11.84GTPGPDD346 pKa = 3.4VMCGGAAADD355 pKa = 4.01SFHH358 pKa = 6.78GAGGEE363 pKa = 4.06DD364 pKa = 3.87IILGGGGNDD373 pKa = 3.38WLNGGTGNDD382 pKa = 3.93TVDD385 pKa = 3.31GGAGQDD391 pKa = 3.51QLRR394 pKa = 11.84GEE396 pKa = 4.63AGADD400 pKa = 3.39ALDD403 pKa = 4.51GGDD406 pKa = 5.18DD407 pKa = 3.71NDD409 pKa = 4.16SVKK412 pKa = 11.04GGVGNDD418 pKa = 3.44TVTGGAGRR426 pKa = 11.84DD427 pKa = 3.68VLDD430 pKa = 3.54EE431 pKa = 4.14TGAAGFGNDD440 pKa = 4.97DD441 pKa = 4.63LDD443 pKa = 5.64GGDD446 pKa = 3.79GNDD449 pKa = 3.62VIRR452 pKa = 11.84GGEE455 pKa = 4.28GVDD458 pKa = 4.41SIAGGDD464 pKa = 3.97GNDD467 pKa = 3.96NLNGGAGSPDD477 pKa = 3.47VCDD480 pKa = 4.63GGPGTDD486 pKa = 2.71IGGYY490 pKa = 9.48GCEE493 pKa = 4.17QKK495 pKa = 11.03VSIPP499 pKa = 3.45
Molecular weight: 48.72 kDa
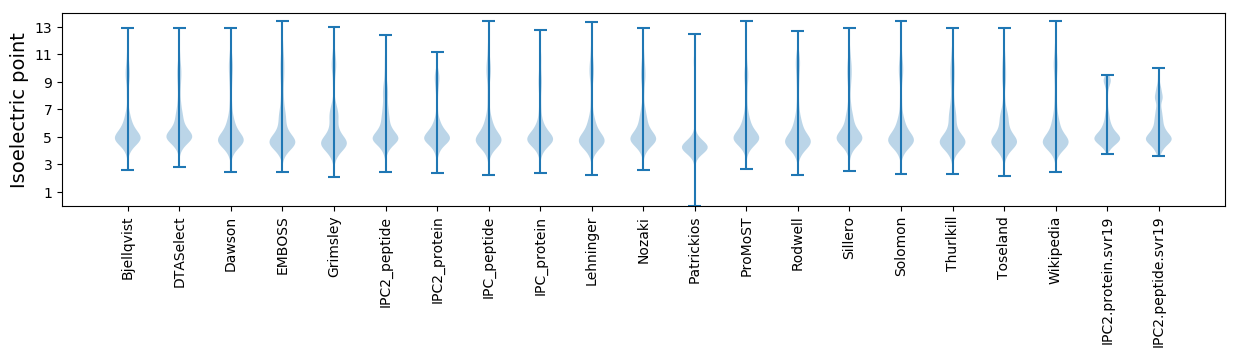
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A316TJC7|A0A316TJC7_9ACTN Respiratory nitrate reductase subunit gamma OS=Nocardioides silvaticus OX=2201891 GN=narI PE=4 SV=1
MM1 pKa = 7.66PGAAGPRR8 pKa = 11.84RR9 pKa = 11.84HH10 pKa = 5.87RR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 5.98RR14 pKa = 11.84RR15 pKa = 11.84PHH17 pKa = 5.98RR18 pKa = 11.84RR19 pKa = 11.84PLLRR23 pKa = 11.84RR24 pKa = 11.84PPPRR28 pKa = 11.84QRR30 pKa = 11.84RR31 pKa = 11.84SRR33 pKa = 11.84RR34 pKa = 11.84TRR36 pKa = 11.84WPRR39 pKa = 11.84STPTSPPASSTRR51 pKa = 11.84RR52 pKa = 11.84AAGGPTTTRR61 pKa = 11.84PPPSRR66 pKa = 11.84SGTRR70 pKa = 11.84AATGSRR76 pKa = 11.84SRR78 pKa = 11.84AVPSCCRR85 pKa = 11.84TSSTRR90 pKa = 11.84SRR92 pKa = 11.84SWATRR97 pKa = 11.84PSSSRR102 pKa = 3.25
MM1 pKa = 7.66PGAAGPRR8 pKa = 11.84RR9 pKa = 11.84HH10 pKa = 5.87RR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 5.98RR14 pKa = 11.84RR15 pKa = 11.84PHH17 pKa = 5.98RR18 pKa = 11.84RR19 pKa = 11.84PLLRR23 pKa = 11.84RR24 pKa = 11.84PPPRR28 pKa = 11.84QRR30 pKa = 11.84RR31 pKa = 11.84SRR33 pKa = 11.84RR34 pKa = 11.84TRR36 pKa = 11.84WPRR39 pKa = 11.84STPTSPPASSTRR51 pKa = 11.84RR52 pKa = 11.84AAGGPTTTRR61 pKa = 11.84PPPSRR66 pKa = 11.84SGTRR70 pKa = 11.84AATGSRR76 pKa = 11.84SRR78 pKa = 11.84AVPSCCRR85 pKa = 11.84TSSTRR90 pKa = 11.84SRR92 pKa = 11.84SWATRR97 pKa = 11.84PSSSRR102 pKa = 3.25
Molecular weight: 11.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1369492 |
33 |
2741 |
324.5 |
34.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.873 ± 0.055 | 0.747 ± 0.01 |
6.783 ± 0.033 | 6.123 ± 0.035 |
2.807 ± 0.021 | 9.177 ± 0.035 |
2.157 ± 0.018 | 3.552 ± 0.028 |
2.067 ± 0.029 | 10.091 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.758 ± 0.015 | 1.79 ± 0.022 |
5.668 ± 0.029 | 2.717 ± 0.019 |
7.628 ± 0.045 | 5.061 ± 0.028 |
6.025 ± 0.034 | 9.38 ± 0.041 |
1.566 ± 0.017 | 2.03 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
