
Lake Sarah-associated circular virus-15
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.63
Get precalculated fractions of proteins
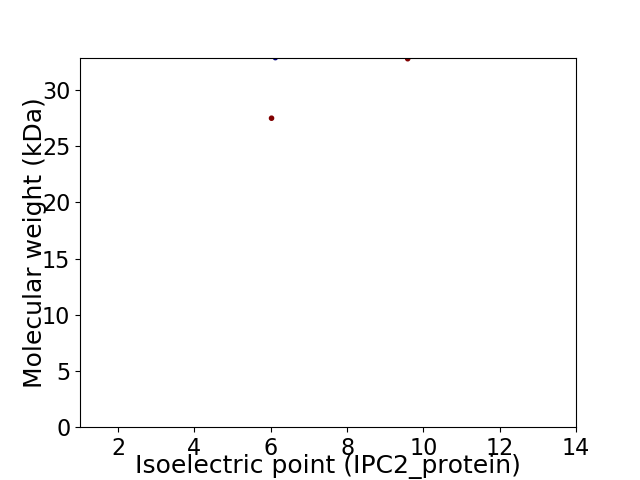
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
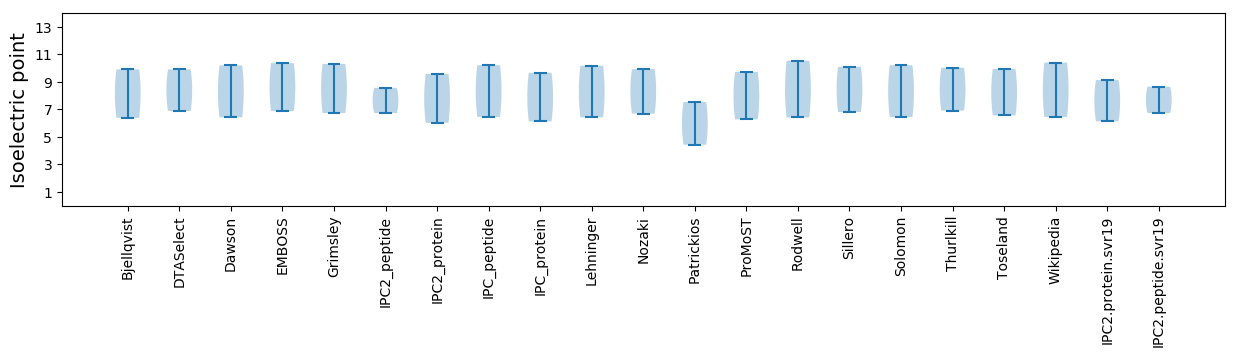
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GA72|A0A126GA72_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-15 OX=1685741 PE=4 SV=1
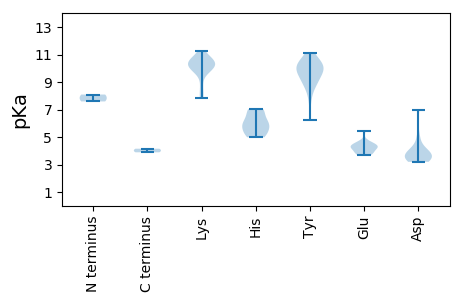
MM1 pKa = 8.09DD2 pKa = 6.98PITDD6 pKa = 3.67TTIKK10 pKa = 7.83STRR13 pKa = 11.84WAFTAYY19 pKa = 9.67EE20 pKa = 4.41GQWHH24 pKa = 7.02LFEE27 pKa = 4.66TMPPEE32 pKa = 3.72IAEE35 pKa = 4.32WGWQTEE41 pKa = 4.27KK42 pKa = 11.28CPDD45 pKa = 3.7TDD47 pKa = 3.37RR48 pKa = 11.84LHH50 pKa = 5.34YY51 pKa = 10.03QGYY54 pKa = 10.89LMLQRR59 pKa = 11.84QQRR62 pKa = 11.84LASLVRR68 pKa = 11.84LLPGVHH74 pKa = 6.83FGAARR79 pKa = 11.84NWTDD83 pKa = 3.3LVNYY87 pKa = 9.05CRR89 pKa = 11.84KK90 pKa = 9.88KK91 pKa = 8.92DD92 pKa = 3.55TRR94 pKa = 11.84VEE96 pKa = 3.96GTEE99 pKa = 3.81QVHH102 pKa = 4.98RR103 pKa = 11.84TNRR106 pKa = 11.84IPTLFGYY113 pKa = 9.71AQEE116 pKa = 4.53VISRR120 pKa = 11.84LPSWDD125 pKa = 4.69EE126 pKa = 3.88IRR128 pKa = 11.84QDD130 pKa = 3.39WITKK134 pKa = 7.92MEE136 pKa = 4.31HH137 pKa = 5.39VSWMLKK143 pKa = 10.02HH144 pKa = 5.92GQPDD148 pKa = 3.56LPDD151 pKa = 3.96PVPRR155 pKa = 11.84SSIFYY160 pKa = 9.74VATSMEE166 pKa = 3.71AHH168 pKa = 7.0AYY170 pKa = 10.42DD171 pKa = 5.12LVTKK175 pKa = 10.62LVGEE179 pKa = 5.42DD180 pKa = 3.23IQKK183 pKa = 10.45GQIGVEE189 pKa = 4.32FIVQNPLWITTFKK202 pKa = 10.69NQIRR206 pKa = 11.84NMVLRR211 pKa = 11.84KK212 pKa = 10.09DD213 pKa = 3.97FQNPPASRR221 pKa = 11.84QTDD224 pKa = 3.48RR225 pKa = 11.84QTEE228 pKa = 3.93ITISFEE234 pKa = 3.89
MM1 pKa = 8.09DD2 pKa = 6.98PITDD6 pKa = 3.67TTIKK10 pKa = 7.83STRR13 pKa = 11.84WAFTAYY19 pKa = 9.67EE20 pKa = 4.41GQWHH24 pKa = 7.02LFEE27 pKa = 4.66TMPPEE32 pKa = 3.72IAEE35 pKa = 4.32WGWQTEE41 pKa = 4.27KK42 pKa = 11.28CPDD45 pKa = 3.7TDD47 pKa = 3.37RR48 pKa = 11.84LHH50 pKa = 5.34YY51 pKa = 10.03QGYY54 pKa = 10.89LMLQRR59 pKa = 11.84QQRR62 pKa = 11.84LASLVRR68 pKa = 11.84LLPGVHH74 pKa = 6.83FGAARR79 pKa = 11.84NWTDD83 pKa = 3.3LVNYY87 pKa = 9.05CRR89 pKa = 11.84KK90 pKa = 9.88KK91 pKa = 8.92DD92 pKa = 3.55TRR94 pKa = 11.84VEE96 pKa = 3.96GTEE99 pKa = 3.81QVHH102 pKa = 4.98RR103 pKa = 11.84TNRR106 pKa = 11.84IPTLFGYY113 pKa = 9.71AQEE116 pKa = 4.53VISRR120 pKa = 11.84LPSWDD125 pKa = 4.69EE126 pKa = 3.88IRR128 pKa = 11.84QDD130 pKa = 3.39WITKK134 pKa = 7.92MEE136 pKa = 4.31HH137 pKa = 5.39VSWMLKK143 pKa = 10.02HH144 pKa = 5.92GQPDD148 pKa = 3.56LPDD151 pKa = 3.96PVPRR155 pKa = 11.84SSIFYY160 pKa = 9.74VATSMEE166 pKa = 3.71AHH168 pKa = 7.0AYY170 pKa = 10.42DD171 pKa = 5.12LVTKK175 pKa = 10.62LVGEE179 pKa = 5.42DD180 pKa = 3.23IQKK183 pKa = 10.45GQIGVEE189 pKa = 4.32FIVQNPLWITTFKK202 pKa = 10.69NQIRR206 pKa = 11.84NMVLRR211 pKa = 11.84KK212 pKa = 10.09DD213 pKa = 3.97FQNPPASRR221 pKa = 11.84QTDD224 pKa = 3.48RR225 pKa = 11.84QTEE228 pKa = 3.93ITISFEE234 pKa = 3.89
Molecular weight: 27.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GA72|A0A126GA72_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-15 OX=1685741 PE=4 SV=1
MM1 pKa = 7.59PPRR4 pKa = 11.84RR5 pKa = 11.84VRR7 pKa = 11.84RR8 pKa = 11.84QRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PARR15 pKa = 11.84KK16 pKa = 8.46YY17 pKa = 7.75RR18 pKa = 11.84QKK20 pKa = 11.02GVRR23 pKa = 11.84TKK25 pKa = 10.64KK26 pKa = 10.18GVKK29 pKa = 9.97ALVTRR34 pKa = 11.84VINAKK39 pKa = 10.62AEE41 pKa = 4.15TKK43 pKa = 9.64MVAFYY48 pKa = 10.48GGPIANPSPLRR59 pKa = 11.84NSTGTYY65 pKa = 10.08ADD67 pKa = 3.96SAPTSQNQYY76 pKa = 9.59ISVNATDD83 pKa = 3.84ILKK86 pKa = 10.18VIPDD90 pKa = 4.0VAPGNADD97 pKa = 3.23NEE99 pKa = 4.38RR100 pKa = 11.84TGRR103 pKa = 11.84YY104 pKa = 9.05INPTSLQLHH113 pKa = 6.32CKK115 pKa = 10.25VGISPISTGGSGYY128 pKa = 10.3QNGWAYY134 pKa = 10.51DD135 pKa = 4.18IIAVAYY141 pKa = 9.2LLQHH145 pKa = 6.08VEE147 pKa = 4.13YY148 pKa = 8.63KK149 pKa = 10.18TYY151 pKa = 10.18QALYY155 pKa = 8.6TNNTFGQLLDD165 pKa = 3.76VMEE168 pKa = 4.65GTTTQFNGDD177 pKa = 3.46FSSANMPVEE186 pKa = 3.72KK187 pKa = 10.21GYY189 pKa = 11.1YY190 pKa = 9.04RR191 pKa = 11.84VLGKK195 pKa = 10.57KK196 pKa = 9.83KK197 pKa = 9.99ISLRR201 pKa = 11.84SSGAYY206 pKa = 9.83NPAAGILNTITNNNSHH222 pKa = 5.79QLSHH226 pKa = 5.89EE227 pKa = 4.2WTWNVGKK234 pKa = 9.66HH235 pKa = 5.06LPKK238 pKa = 10.49KK239 pKa = 10.47LIYY242 pKa = 10.07PEE244 pKa = 4.3GTVSVANGQNEE255 pKa = 4.55PLNAAPFWAVGYY267 pKa = 9.9YY268 pKa = 10.41NVDD271 pKa = 3.2GTGGSPATAKK281 pKa = 10.53INIQQQYY288 pKa = 6.23TAIMKK293 pKa = 10.38FKK295 pKa = 10.97DD296 pKa = 3.51FF297 pKa = 4.15
MM1 pKa = 7.59PPRR4 pKa = 11.84RR5 pKa = 11.84VRR7 pKa = 11.84RR8 pKa = 11.84QRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PARR15 pKa = 11.84KK16 pKa = 8.46YY17 pKa = 7.75RR18 pKa = 11.84QKK20 pKa = 11.02GVRR23 pKa = 11.84TKK25 pKa = 10.64KK26 pKa = 10.18GVKK29 pKa = 9.97ALVTRR34 pKa = 11.84VINAKK39 pKa = 10.62AEE41 pKa = 4.15TKK43 pKa = 9.64MVAFYY48 pKa = 10.48GGPIANPSPLRR59 pKa = 11.84NSTGTYY65 pKa = 10.08ADD67 pKa = 3.96SAPTSQNQYY76 pKa = 9.59ISVNATDD83 pKa = 3.84ILKK86 pKa = 10.18VIPDD90 pKa = 4.0VAPGNADD97 pKa = 3.23NEE99 pKa = 4.38RR100 pKa = 11.84TGRR103 pKa = 11.84YY104 pKa = 9.05INPTSLQLHH113 pKa = 6.32CKK115 pKa = 10.25VGISPISTGGSGYY128 pKa = 10.3QNGWAYY134 pKa = 10.51DD135 pKa = 4.18IIAVAYY141 pKa = 9.2LLQHH145 pKa = 6.08VEE147 pKa = 4.13YY148 pKa = 8.63KK149 pKa = 10.18TYY151 pKa = 10.18QALYY155 pKa = 8.6TNNTFGQLLDD165 pKa = 3.76VMEE168 pKa = 4.65GTTTQFNGDD177 pKa = 3.46FSSANMPVEE186 pKa = 3.72KK187 pKa = 10.21GYY189 pKa = 11.1YY190 pKa = 9.04RR191 pKa = 11.84VLGKK195 pKa = 10.57KK196 pKa = 9.83KK197 pKa = 9.99ISLRR201 pKa = 11.84SSGAYY206 pKa = 9.83NPAAGILNTITNNNSHH222 pKa = 5.79QLSHH226 pKa = 5.89EE227 pKa = 4.2WTWNVGKK234 pKa = 9.66HH235 pKa = 5.06LPKK238 pKa = 10.49KK239 pKa = 10.47LIYY242 pKa = 10.07PEE244 pKa = 4.3GTVSVANGQNEE255 pKa = 4.55PLNAAPFWAVGYY267 pKa = 9.9YY268 pKa = 10.41NVDD271 pKa = 3.2GTGGSPATAKK281 pKa = 10.53INIQQQYY288 pKa = 6.23TAIMKK293 pKa = 10.38FKK295 pKa = 10.97DD296 pKa = 3.51FF297 pKa = 4.15
Molecular weight: 32.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
531 |
234 |
297 |
265.5 |
30.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.968 ± 1.458 | 0.565 ± 0.186 |
4.331 ± 1.062 | 4.331 ± 1.337 |
3.013 ± 0.536 | 6.968 ± 1.458 |
2.26 ± 0.47 | 6.026 ± 0.247 |
5.65 ± 0.885 | 6.403 ± 0.554 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.26 ± 0.47 | 5.838 ± 1.83 |
6.026 ± 0.028 | 6.026 ± 0.796 |
6.215 ± 0.675 | 5.273 ± 0.643 |
8.286 ± 0.442 | 6.403 ± 0.27 |
2.448 ± 0.899 | 4.708 ± 1.104 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
