
Thermomonas fusca
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Thermomonas
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
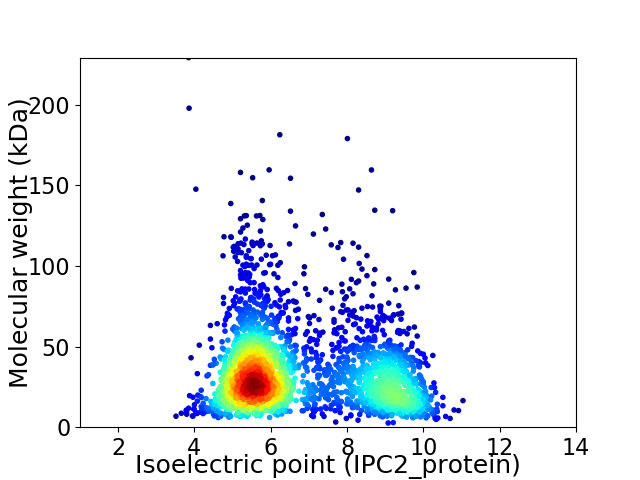
Virtual 2D-PAGE plot for 2622 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
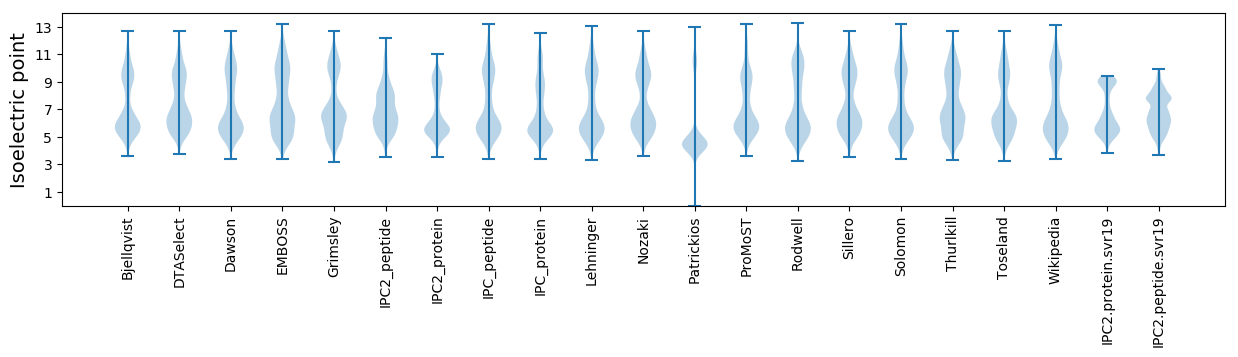
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5R9PIB8|A0A5R9PIB8_9GAMM Ferredoxin--NADP reductase OS=Thermomonas fusca OX=215690 GN=E5S66_01420 PE=3 SV=1
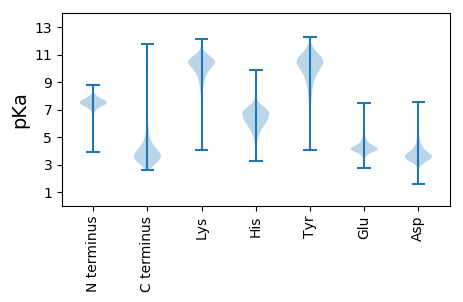
MM1 pKa = 7.97SEE3 pKa = 3.33IRR5 pKa = 11.84IVGKK9 pKa = 9.49PGKK12 pKa = 8.97ATRR15 pKa = 11.84KK16 pKa = 8.75PRR18 pKa = 11.84TVKK21 pKa = 10.16LAPMARR27 pKa = 11.84AVRR30 pKa = 11.84SALAVSAMAFALGTTGNVMAGAHH53 pKa = 5.25EE54 pKa = 4.7VSTAHH59 pKa = 7.28ALQIQRR65 pKa = 11.84AAIDD69 pKa = 3.71FAPVFDD75 pKa = 4.3LTVVGAGPVALAIDD89 pKa = 3.78VTDD92 pKa = 4.73PGDD95 pKa = 3.56IVIDD99 pKa = 3.69NVDD102 pKa = 4.6PISEE106 pKa = 4.28YY107 pKa = 11.18AAGDD111 pKa = 4.26AIAIRR116 pKa = 11.84GYY118 pKa = 8.87STGGNVDD125 pKa = 2.96ITNAATGVLIADD137 pKa = 4.27AGYY140 pKa = 10.61GNAIGIYY147 pKa = 9.34GYY149 pKa = 10.66SATGDD154 pKa = 3.51VAINNAGDD162 pKa = 3.28ITAYY166 pKa = 10.21SYY168 pKa = 11.53SGLADD173 pKa = 4.9GIFASGDD180 pKa = 3.47HH181 pKa = 5.61VVVDD185 pKa = 3.39NSGAIDD191 pKa = 3.82AYY193 pKa = 10.5GYY195 pKa = 10.8AWAAGIEE202 pKa = 4.05ARR204 pKa = 11.84ANTSVEE210 pKa = 4.14VTNSGDD216 pKa = 3.06IYY218 pKa = 11.06AGIYY222 pKa = 8.15ATGGAYY228 pKa = 10.11GIYY231 pKa = 10.38ASGHH235 pKa = 6.46DD236 pKa = 3.57ATVVNDD242 pKa = 3.82GGITARR248 pKa = 11.84GYY250 pKa = 8.91YY251 pKa = 8.81ATGIRR256 pKa = 11.84TQTYY260 pKa = 8.41GDD262 pKa = 3.63AVIDD266 pKa = 3.54NTGNIDD272 pKa = 3.34AGTTTGSALAWGINAVASGEE292 pKa = 4.11GAAIQIDD299 pKa = 3.93NTGSIQAAAVYY310 pKa = 9.49GATGIEE316 pKa = 4.82GIATGTGGTVQVNNSGDD333 pKa = 3.14IVVQQNRR340 pKa = 11.84AITSYY345 pKa = 10.89GGYY348 pKa = 10.38GILASADD355 pKa = 2.94GDD357 pKa = 3.79ASINNSGDD365 pKa = 3.0IVVASKK371 pKa = 11.19GPAFGLVGMSFSGEE385 pKa = 3.32ASVTNEE391 pKa = 3.4GNVAVSSTVNSTGVTPTGLLAFGANGTAAVEE422 pKa = 4.02NSGTVVATGVRR433 pKa = 11.84RR434 pKa = 11.84AAGIDD439 pKa = 3.03ASAYY443 pKa = 10.17GDD445 pKa = 3.56VSIDD449 pKa = 3.37NSGDD453 pKa = 3.0VSAYY457 pKa = 7.78ATGMFTNTRR466 pKa = 11.84ADD468 pKa = 3.91GIKK471 pKa = 10.15AVSGSGDD478 pKa = 2.98ITLDD482 pKa = 3.44NSGSIYY488 pKa = 10.8ASGTTVGLGVYY499 pKa = 10.06AVAEE503 pKa = 4.14NGNIAINNAVDD514 pKa = 4.02GSIGAYY520 pKa = 9.61SYY522 pKa = 11.16YY523 pKa = 10.65GVARR527 pKa = 11.84GVFAYY532 pKa = 7.75ATHH535 pKa = 6.85GDD537 pKa = 3.42IAIDD541 pKa = 3.41SAGAIDD547 pKa = 4.06AYY549 pKa = 10.63GWTRR553 pKa = 11.84TYY555 pKa = 11.38GVMALAPGGDD565 pKa = 3.54AAVDD569 pKa = 3.66SSGTIYY575 pKa = 10.85ARR577 pKa = 11.84SGSNNATGVLASTDD591 pKa = 3.21QGAASITNSGDD602 pKa = 2.95IQVISRR608 pKa = 11.84TGSYY612 pKa = 10.17GVRR615 pKa = 11.84AYY617 pKa = 10.27GYY619 pKa = 10.87AGADD623 pKa = 3.42VGNSGSIYY631 pKa = 9.99AASTNGVAVGMMAQAGDD648 pKa = 3.68GNATVTNTGGSIEE661 pKa = 4.31VKK663 pKa = 10.33AATAAFGIFADD674 pKa = 4.15SAYY677 pKa = 10.86GDD679 pKa = 3.57ATVTNASEE687 pKa = 4.06ISALGVNSAATGIRR701 pKa = 11.84ATASGDD707 pKa = 3.29VDD709 pKa = 3.38ITNSATIDD717 pKa = 3.3AMSNGDD723 pKa = 3.76AIGIYY728 pKa = 10.01GYY730 pKa = 11.0SLAADD735 pKa = 3.56VTIDD739 pKa = 3.39NSGDD743 pKa = 2.99ITASSSYY750 pKa = 10.83GLADD754 pKa = 4.96GIFASGQDD762 pKa = 3.49VAVTNSGAVSVTSVYY777 pKa = 10.96GDD779 pKa = 3.2WAAGIEE785 pKa = 4.29AQGSNSTQVSNTGDD799 pKa = 3.09ITASAMPFLQVEE811 pKa = 4.67DD812 pKa = 4.26GTSVLYY818 pKa = 9.5STAGGQAFGIYY829 pKa = 8.51ATAGVGNGVVTNSGSLVVQGGYY851 pKa = 8.63ATGIEE856 pKa = 4.14AQSEE860 pKa = 4.39GDD862 pKa = 3.53LSVTNSGDD870 pKa = 2.98IFAGTGLGAYY880 pKa = 9.32YY881 pKa = 10.53NSNNGYY887 pKa = 9.3GYY889 pKa = 11.1YY890 pKa = 10.22YY891 pKa = 9.03GTLMAIGINASGNGEE906 pKa = 4.03GMHH909 pKa = 5.83VAVANTGDD917 pKa = 3.26IAANGIFGASGISATASGLGGTAVVTNTGDD947 pKa = 2.84ITAAQYY953 pKa = 10.56VAGGYY958 pKa = 9.7GAFGIVASTDD968 pKa = 3.18GDD970 pKa = 3.87SGIDD974 pKa = 3.24NGGAITVTSGGTGYY988 pKa = 10.9GVAALSFAGDD998 pKa = 3.3AVVVNSGDD1006 pKa = 3.58VSVEE1010 pKa = 3.52ASTAASKK1017 pKa = 11.3YY1018 pKa = 7.9YY1019 pKa = 10.8GSYY1022 pKa = 10.93GLLAFAGNGYY1032 pKa = 10.19AAVDD1036 pKa = 3.54NSGSVSVSNAGIMAAPARR1054 pKa = 11.84AVDD1057 pKa = 3.54AQGQQGVGVVNSGALYY1073 pKa = 10.82ADD1075 pKa = 3.73GKK1077 pKa = 9.16YY1078 pKa = 10.48AYY1080 pKa = 10.15GVFAVSAGGDD1090 pKa = 3.36VAVNNTADD1098 pKa = 3.75GDD1100 pKa = 3.81IGFYY1104 pKa = 10.64SYY1106 pKa = 11.18LGYY1109 pKa = 8.98GTGILGVSEE1118 pKa = 4.75GGDD1121 pKa = 3.41VAIGNAGAIAGYY1133 pKa = 9.92GYY1135 pKa = 10.17SQAAGIFGGASTGDD1149 pKa = 3.14VDD1151 pKa = 4.92VSNASTGSIDD1161 pKa = 3.56VVSAGFAVGIFARR1174 pKa = 11.84ADD1176 pKa = 3.29YY1177 pKa = 8.41GTATIVNSGDD1187 pKa = 2.73ISASVFPDD1195 pKa = 3.01TGYY1198 pKa = 10.88QGDD1201 pKa = 3.64VAYY1204 pKa = 10.43GLLATSDD1211 pKa = 3.7YY1212 pKa = 11.14GYY1214 pKa = 11.16AAAGNSGDD1222 pKa = 3.67IQADD1226 pKa = 4.44GYY1228 pKa = 9.19YY1229 pKa = 9.28TATGIAARR1237 pKa = 11.84SLLGSTVSTAAGSHH1251 pKa = 5.9INATADD1257 pKa = 3.38YY1258 pKa = 9.58VAIGIEE1264 pKa = 4.28GRR1266 pKa = 11.84AEE1268 pKa = 4.25GGDD1271 pKa = 3.54VTVGSAGDD1279 pKa = 3.25IATAGGYY1286 pKa = 10.49GSVGIQAYY1294 pKa = 10.35SAMGDD1299 pKa = 3.42VTVGNTGHH1307 pKa = 6.29ITAASGYY1314 pKa = 9.14GIAIGATAYY1323 pKa = 9.74TMEE1326 pKa = 4.38GLVTVTNNGLIEE1338 pKa = 4.72ADD1340 pKa = 3.53AAIASYY1346 pKa = 10.7GILADD1351 pKa = 4.06SYY1353 pKa = 11.78AGDD1356 pKa = 3.51VVVNLGSNGRR1366 pKa = 11.84VEE1368 pKa = 5.16ASAGMAAFGVVVNSGDD1384 pKa = 3.67GDD1386 pKa = 4.04STVNNAGKK1394 pKa = 9.35IHH1396 pKa = 6.99AGGATYY1402 pKa = 11.03NFGVVFEE1409 pKa = 4.58GMYY1412 pKa = 10.48GANTLNNLATGVISADD1428 pKa = 3.21GDD1430 pKa = 3.42EE1431 pKa = 5.23GYY1433 pKa = 11.51AFAVAGNDD1441 pKa = 4.2GVEE1444 pKa = 4.18TLNNSGRR1451 pKa = 11.84ILGAISLYY1459 pKa = 11.1GGDD1462 pKa = 4.56DD1463 pKa = 3.58VFSNKK1468 pKa = 10.09AGGTWDD1474 pKa = 3.33VGSTLSTDD1482 pKa = 4.17FGYY1485 pKa = 11.44GNDD1488 pKa = 3.73TLANAAGGTITLNGGQIAFGAGDD1511 pKa = 3.69DD1512 pKa = 4.32TINNAGLIGLTNGTITMGDD1531 pKa = 3.59ASPVVPLQAAAPQAVEE1547 pKa = 3.94MNAFNNSGTLKK1558 pKa = 10.29IVGSSVVDD1566 pKa = 3.29TAGGVFTNTGLVDD1579 pKa = 4.05FVNGVTTDD1587 pKa = 3.43ALSLDD1592 pKa = 3.65GTLAGSGQMAIDD1604 pKa = 3.73VNFNDD1609 pKa = 4.07LTADD1613 pKa = 3.39QLAINGDD1620 pKa = 3.7IASGTAQTVNVKK1632 pKa = 10.34FIGVPTLGAPAVEE1645 pKa = 4.62FASVTGTSAAGSFVGGHH1662 pKa = 4.25VLGYY1666 pKa = 9.47NQNANFLTLGVDD1678 pKa = 3.49VTSVLNAGNAADD1690 pKa = 4.75DD1691 pKa = 4.28LFSVGLRR1698 pKa = 11.84VDD1700 pKa = 3.72GLSDD1704 pKa = 4.14GGTLAANAASGAAGFMNSQVGTFRR1728 pKa = 11.84QRR1730 pKa = 11.84LGVNPYY1736 pKa = 10.14GDD1738 pKa = 3.57AGKK1741 pKa = 10.48VLSAFVRR1748 pKa = 11.84VYY1750 pKa = 10.62TDD1752 pKa = 3.0QGDD1755 pKa = 3.89AKK1757 pKa = 9.06PTHH1760 pKa = 6.26LAGNFGQGGNFAYY1773 pKa = 9.81EE1774 pKa = 3.73QASWGRR1780 pKa = 11.84EE1781 pKa = 3.54FGVNANLFGNLHH1793 pKa = 6.66AGVVMGNAASRR1804 pKa = 11.84QRR1806 pKa = 11.84LTDD1809 pKa = 3.44GEE1811 pKa = 4.56GQNRR1815 pKa = 11.84MNGTTFGAYY1824 pKa = 7.35ATWYY1828 pKa = 9.34VPEE1831 pKa = 4.35GLYY1834 pKa = 10.64VDD1836 pKa = 5.31LSGRR1840 pKa = 11.84WMGADD1845 pKa = 2.93IRR1847 pKa = 11.84SDD1849 pKa = 3.53SVGANLQSRR1858 pKa = 11.84ARR1860 pKa = 11.84TSAVSLEE1867 pKa = 4.69AGYY1870 pKa = 9.59EE1871 pKa = 3.97WKK1873 pKa = 10.6LGSVNVVPQAQYY1885 pKa = 10.49IRR1887 pKa = 11.84TRR1889 pKa = 11.84VEE1891 pKa = 3.89DD1892 pKa = 3.08VRR1894 pKa = 11.84TFYY1897 pKa = 11.39GEE1899 pKa = 4.19SLDD1902 pKa = 3.71FTVQGGTFEE1911 pKa = 4.2RR1912 pKa = 11.84GRR1914 pKa = 11.84VGVEE1918 pKa = 3.66VNKK1921 pKa = 9.4TFQSGDD1927 pKa = 3.54IRR1929 pKa = 11.84WTPYY1933 pKa = 10.85GSISALRR1940 pKa = 11.84DD1941 pKa = 3.41SGGKK1945 pKa = 7.85TSYY1948 pKa = 11.24AVGDD1952 pKa = 4.23FYY1954 pKa = 11.73GSTGVSGSRR1963 pKa = 11.84AAAEE1967 pKa = 4.32LGVGVQKK1974 pKa = 11.1GGLGVTLGVNWLDD1987 pKa = 3.74GGSLKK1992 pKa = 10.99SFIGGQANLRR2002 pKa = 11.84YY2003 pKa = 9.59SWW2005 pKa = 3.2
MM1 pKa = 7.97SEE3 pKa = 3.33IRR5 pKa = 11.84IVGKK9 pKa = 9.49PGKK12 pKa = 8.97ATRR15 pKa = 11.84KK16 pKa = 8.75PRR18 pKa = 11.84TVKK21 pKa = 10.16LAPMARR27 pKa = 11.84AVRR30 pKa = 11.84SALAVSAMAFALGTTGNVMAGAHH53 pKa = 5.25EE54 pKa = 4.7VSTAHH59 pKa = 7.28ALQIQRR65 pKa = 11.84AAIDD69 pKa = 3.71FAPVFDD75 pKa = 4.3LTVVGAGPVALAIDD89 pKa = 3.78VTDD92 pKa = 4.73PGDD95 pKa = 3.56IVIDD99 pKa = 3.69NVDD102 pKa = 4.6PISEE106 pKa = 4.28YY107 pKa = 11.18AAGDD111 pKa = 4.26AIAIRR116 pKa = 11.84GYY118 pKa = 8.87STGGNVDD125 pKa = 2.96ITNAATGVLIADD137 pKa = 4.27AGYY140 pKa = 10.61GNAIGIYY147 pKa = 9.34GYY149 pKa = 10.66SATGDD154 pKa = 3.51VAINNAGDD162 pKa = 3.28ITAYY166 pKa = 10.21SYY168 pKa = 11.53SGLADD173 pKa = 4.9GIFASGDD180 pKa = 3.47HH181 pKa = 5.61VVVDD185 pKa = 3.39NSGAIDD191 pKa = 3.82AYY193 pKa = 10.5GYY195 pKa = 10.8AWAAGIEE202 pKa = 4.05ARR204 pKa = 11.84ANTSVEE210 pKa = 4.14VTNSGDD216 pKa = 3.06IYY218 pKa = 11.06AGIYY222 pKa = 8.15ATGGAYY228 pKa = 10.11GIYY231 pKa = 10.38ASGHH235 pKa = 6.46DD236 pKa = 3.57ATVVNDD242 pKa = 3.82GGITARR248 pKa = 11.84GYY250 pKa = 8.91YY251 pKa = 8.81ATGIRR256 pKa = 11.84TQTYY260 pKa = 8.41GDD262 pKa = 3.63AVIDD266 pKa = 3.54NTGNIDD272 pKa = 3.34AGTTTGSALAWGINAVASGEE292 pKa = 4.11GAAIQIDD299 pKa = 3.93NTGSIQAAAVYY310 pKa = 9.49GATGIEE316 pKa = 4.82GIATGTGGTVQVNNSGDD333 pKa = 3.14IVVQQNRR340 pKa = 11.84AITSYY345 pKa = 10.89GGYY348 pKa = 10.38GILASADD355 pKa = 2.94GDD357 pKa = 3.79ASINNSGDD365 pKa = 3.0IVVASKK371 pKa = 11.19GPAFGLVGMSFSGEE385 pKa = 3.32ASVTNEE391 pKa = 3.4GNVAVSSTVNSTGVTPTGLLAFGANGTAAVEE422 pKa = 4.02NSGTVVATGVRR433 pKa = 11.84RR434 pKa = 11.84AAGIDD439 pKa = 3.03ASAYY443 pKa = 10.17GDD445 pKa = 3.56VSIDD449 pKa = 3.37NSGDD453 pKa = 3.0VSAYY457 pKa = 7.78ATGMFTNTRR466 pKa = 11.84ADD468 pKa = 3.91GIKK471 pKa = 10.15AVSGSGDD478 pKa = 2.98ITLDD482 pKa = 3.44NSGSIYY488 pKa = 10.8ASGTTVGLGVYY499 pKa = 10.06AVAEE503 pKa = 4.14NGNIAINNAVDD514 pKa = 4.02GSIGAYY520 pKa = 9.61SYY522 pKa = 11.16YY523 pKa = 10.65GVARR527 pKa = 11.84GVFAYY532 pKa = 7.75ATHH535 pKa = 6.85GDD537 pKa = 3.42IAIDD541 pKa = 3.41SAGAIDD547 pKa = 4.06AYY549 pKa = 10.63GWTRR553 pKa = 11.84TYY555 pKa = 11.38GVMALAPGGDD565 pKa = 3.54AAVDD569 pKa = 3.66SSGTIYY575 pKa = 10.85ARR577 pKa = 11.84SGSNNATGVLASTDD591 pKa = 3.21QGAASITNSGDD602 pKa = 2.95IQVISRR608 pKa = 11.84TGSYY612 pKa = 10.17GVRR615 pKa = 11.84AYY617 pKa = 10.27GYY619 pKa = 10.87AGADD623 pKa = 3.42VGNSGSIYY631 pKa = 9.99AASTNGVAVGMMAQAGDD648 pKa = 3.68GNATVTNTGGSIEE661 pKa = 4.31VKK663 pKa = 10.33AATAAFGIFADD674 pKa = 4.15SAYY677 pKa = 10.86GDD679 pKa = 3.57ATVTNASEE687 pKa = 4.06ISALGVNSAATGIRR701 pKa = 11.84ATASGDD707 pKa = 3.29VDD709 pKa = 3.38ITNSATIDD717 pKa = 3.3AMSNGDD723 pKa = 3.76AIGIYY728 pKa = 10.01GYY730 pKa = 11.0SLAADD735 pKa = 3.56VTIDD739 pKa = 3.39NSGDD743 pKa = 2.99ITASSSYY750 pKa = 10.83GLADD754 pKa = 4.96GIFASGQDD762 pKa = 3.49VAVTNSGAVSVTSVYY777 pKa = 10.96GDD779 pKa = 3.2WAAGIEE785 pKa = 4.29AQGSNSTQVSNTGDD799 pKa = 3.09ITASAMPFLQVEE811 pKa = 4.67DD812 pKa = 4.26GTSVLYY818 pKa = 9.5STAGGQAFGIYY829 pKa = 8.51ATAGVGNGVVTNSGSLVVQGGYY851 pKa = 8.63ATGIEE856 pKa = 4.14AQSEE860 pKa = 4.39GDD862 pKa = 3.53LSVTNSGDD870 pKa = 2.98IFAGTGLGAYY880 pKa = 9.32YY881 pKa = 10.53NSNNGYY887 pKa = 9.3GYY889 pKa = 11.1YY890 pKa = 10.22YY891 pKa = 9.03GTLMAIGINASGNGEE906 pKa = 4.03GMHH909 pKa = 5.83VAVANTGDD917 pKa = 3.26IAANGIFGASGISATASGLGGTAVVTNTGDD947 pKa = 2.84ITAAQYY953 pKa = 10.56VAGGYY958 pKa = 9.7GAFGIVASTDD968 pKa = 3.18GDD970 pKa = 3.87SGIDD974 pKa = 3.24NGGAITVTSGGTGYY988 pKa = 10.9GVAALSFAGDD998 pKa = 3.3AVVVNSGDD1006 pKa = 3.58VSVEE1010 pKa = 3.52ASTAASKK1017 pKa = 11.3YY1018 pKa = 7.9YY1019 pKa = 10.8GSYY1022 pKa = 10.93GLLAFAGNGYY1032 pKa = 10.19AAVDD1036 pKa = 3.54NSGSVSVSNAGIMAAPARR1054 pKa = 11.84AVDD1057 pKa = 3.54AQGQQGVGVVNSGALYY1073 pKa = 10.82ADD1075 pKa = 3.73GKK1077 pKa = 9.16YY1078 pKa = 10.48AYY1080 pKa = 10.15GVFAVSAGGDD1090 pKa = 3.36VAVNNTADD1098 pKa = 3.75GDD1100 pKa = 3.81IGFYY1104 pKa = 10.64SYY1106 pKa = 11.18LGYY1109 pKa = 8.98GTGILGVSEE1118 pKa = 4.75GGDD1121 pKa = 3.41VAIGNAGAIAGYY1133 pKa = 9.92GYY1135 pKa = 10.17SQAAGIFGGASTGDD1149 pKa = 3.14VDD1151 pKa = 4.92VSNASTGSIDD1161 pKa = 3.56VVSAGFAVGIFARR1174 pKa = 11.84ADD1176 pKa = 3.29YY1177 pKa = 8.41GTATIVNSGDD1187 pKa = 2.73ISASVFPDD1195 pKa = 3.01TGYY1198 pKa = 10.88QGDD1201 pKa = 3.64VAYY1204 pKa = 10.43GLLATSDD1211 pKa = 3.7YY1212 pKa = 11.14GYY1214 pKa = 11.16AAAGNSGDD1222 pKa = 3.67IQADD1226 pKa = 4.44GYY1228 pKa = 9.19YY1229 pKa = 9.28TATGIAARR1237 pKa = 11.84SLLGSTVSTAAGSHH1251 pKa = 5.9INATADD1257 pKa = 3.38YY1258 pKa = 9.58VAIGIEE1264 pKa = 4.28GRR1266 pKa = 11.84AEE1268 pKa = 4.25GGDD1271 pKa = 3.54VTVGSAGDD1279 pKa = 3.25IATAGGYY1286 pKa = 10.49GSVGIQAYY1294 pKa = 10.35SAMGDD1299 pKa = 3.42VTVGNTGHH1307 pKa = 6.29ITAASGYY1314 pKa = 9.14GIAIGATAYY1323 pKa = 9.74TMEE1326 pKa = 4.38GLVTVTNNGLIEE1338 pKa = 4.72ADD1340 pKa = 3.53AAIASYY1346 pKa = 10.7GILADD1351 pKa = 4.06SYY1353 pKa = 11.78AGDD1356 pKa = 3.51VVVNLGSNGRR1366 pKa = 11.84VEE1368 pKa = 5.16ASAGMAAFGVVVNSGDD1384 pKa = 3.67GDD1386 pKa = 4.04STVNNAGKK1394 pKa = 9.35IHH1396 pKa = 6.99AGGATYY1402 pKa = 11.03NFGVVFEE1409 pKa = 4.58GMYY1412 pKa = 10.48GANTLNNLATGVISADD1428 pKa = 3.21GDD1430 pKa = 3.42EE1431 pKa = 5.23GYY1433 pKa = 11.51AFAVAGNDD1441 pKa = 4.2GVEE1444 pKa = 4.18TLNNSGRR1451 pKa = 11.84ILGAISLYY1459 pKa = 11.1GGDD1462 pKa = 4.56DD1463 pKa = 3.58VFSNKK1468 pKa = 10.09AGGTWDD1474 pKa = 3.33VGSTLSTDD1482 pKa = 4.17FGYY1485 pKa = 11.44GNDD1488 pKa = 3.73TLANAAGGTITLNGGQIAFGAGDD1511 pKa = 3.69DD1512 pKa = 4.32TINNAGLIGLTNGTITMGDD1531 pKa = 3.59ASPVVPLQAAAPQAVEE1547 pKa = 3.94MNAFNNSGTLKK1558 pKa = 10.29IVGSSVVDD1566 pKa = 3.29TAGGVFTNTGLVDD1579 pKa = 4.05FVNGVTTDD1587 pKa = 3.43ALSLDD1592 pKa = 3.65GTLAGSGQMAIDD1604 pKa = 3.73VNFNDD1609 pKa = 4.07LTADD1613 pKa = 3.39QLAINGDD1620 pKa = 3.7IASGTAQTVNVKK1632 pKa = 10.34FIGVPTLGAPAVEE1645 pKa = 4.62FASVTGTSAAGSFVGGHH1662 pKa = 4.25VLGYY1666 pKa = 9.47NQNANFLTLGVDD1678 pKa = 3.49VTSVLNAGNAADD1690 pKa = 4.75DD1691 pKa = 4.28LFSVGLRR1698 pKa = 11.84VDD1700 pKa = 3.72GLSDD1704 pKa = 4.14GGTLAANAASGAAGFMNSQVGTFRR1728 pKa = 11.84QRR1730 pKa = 11.84LGVNPYY1736 pKa = 10.14GDD1738 pKa = 3.57AGKK1741 pKa = 10.48VLSAFVRR1748 pKa = 11.84VYY1750 pKa = 10.62TDD1752 pKa = 3.0QGDD1755 pKa = 3.89AKK1757 pKa = 9.06PTHH1760 pKa = 6.26LAGNFGQGGNFAYY1773 pKa = 9.81EE1774 pKa = 3.73QASWGRR1780 pKa = 11.84EE1781 pKa = 3.54FGVNANLFGNLHH1793 pKa = 6.66AGVVMGNAASRR1804 pKa = 11.84QRR1806 pKa = 11.84LTDD1809 pKa = 3.44GEE1811 pKa = 4.56GQNRR1815 pKa = 11.84MNGTTFGAYY1824 pKa = 7.35ATWYY1828 pKa = 9.34VPEE1831 pKa = 4.35GLYY1834 pKa = 10.64VDD1836 pKa = 5.31LSGRR1840 pKa = 11.84WMGADD1845 pKa = 2.93IRR1847 pKa = 11.84SDD1849 pKa = 3.53SVGANLQSRR1858 pKa = 11.84ARR1860 pKa = 11.84TSAVSLEE1867 pKa = 4.69AGYY1870 pKa = 9.59EE1871 pKa = 3.97WKK1873 pKa = 10.6LGSVNVVPQAQYY1885 pKa = 10.49IRR1887 pKa = 11.84TRR1889 pKa = 11.84VEE1891 pKa = 3.89DD1892 pKa = 3.08VRR1894 pKa = 11.84TFYY1897 pKa = 11.39GEE1899 pKa = 4.19SLDD1902 pKa = 3.71FTVQGGTFEE1911 pKa = 4.2RR1912 pKa = 11.84GRR1914 pKa = 11.84VGVEE1918 pKa = 3.66VNKK1921 pKa = 9.4TFQSGDD1927 pKa = 3.54IRR1929 pKa = 11.84WTPYY1933 pKa = 10.85GSISALRR1940 pKa = 11.84DD1941 pKa = 3.41SGGKK1945 pKa = 7.85TSYY1948 pKa = 11.24AVGDD1952 pKa = 4.23FYY1954 pKa = 11.73GSTGVSGSRR1963 pKa = 11.84AAAEE1967 pKa = 4.32LGVGVQKK1974 pKa = 11.1GGLGVTLGVNWLDD1987 pKa = 3.74GGSLKK1992 pKa = 10.99SFIGGQANLRR2002 pKa = 11.84YY2003 pKa = 9.59SWW2005 pKa = 3.2
Molecular weight: 197.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5R9PHK0|A0A5R9PHK0_9GAMM NrdJb OS=Thermomonas fusca OX=215690 GN=E5S66_02270 PE=4 SV=1
MM1 pKa = 6.72THH3 pKa = 7.26RR4 pKa = 11.84DD5 pKa = 3.18LDD7 pKa = 3.73ALLRR11 pKa = 11.84QLVILGALAVLLIPAARR28 pKa = 11.84GSSALLGWLPLWLLGMPLAAWWSLRR53 pKa = 11.84RR54 pKa = 11.84FPLPTLALRR63 pKa = 11.84LPQRR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84VQARR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84GHH78 pKa = 5.53RR79 pKa = 11.84VARR82 pKa = 11.84ALRR85 pKa = 11.84IARR88 pKa = 11.84RR89 pKa = 11.84AA90 pKa = 3.26
MM1 pKa = 6.72THH3 pKa = 7.26RR4 pKa = 11.84DD5 pKa = 3.18LDD7 pKa = 3.73ALLRR11 pKa = 11.84QLVILGALAVLLIPAARR28 pKa = 11.84GSSALLGWLPLWLLGMPLAAWWSLRR53 pKa = 11.84RR54 pKa = 11.84FPLPTLALRR63 pKa = 11.84LPQRR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84VQARR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84GHH78 pKa = 5.53RR79 pKa = 11.84VARR82 pKa = 11.84ALRR85 pKa = 11.84IARR88 pKa = 11.84RR89 pKa = 11.84AA90 pKa = 3.26
Molecular weight: 10.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
884264 |
25 |
2306 |
337.2 |
36.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.912 ± 0.072 | 0.84 ± 0.013 |
5.866 ± 0.033 | 5.294 ± 0.038 |
3.325 ± 0.029 | 8.692 ± 0.05 |
2.2 ± 0.024 | 4.086 ± 0.035 |
2.855 ± 0.043 | 11.153 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.249 ± 0.023 | 2.385 ± 0.029 |
5.257 ± 0.033 | 3.753 ± 0.031 |
7.803 ± 0.052 | 4.89 ± 0.038 |
4.523 ± 0.043 | 7.256 ± 0.039 |
1.492 ± 0.021 | 2.166 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
