
Circovirus-like genome DCCV-2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.23
Get precalculated fractions of proteins
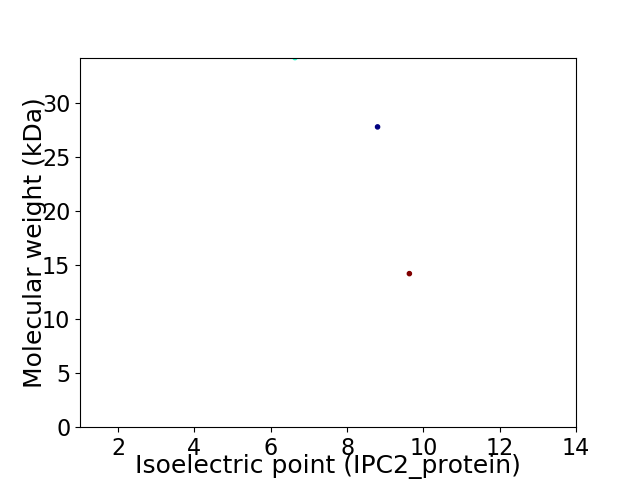
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
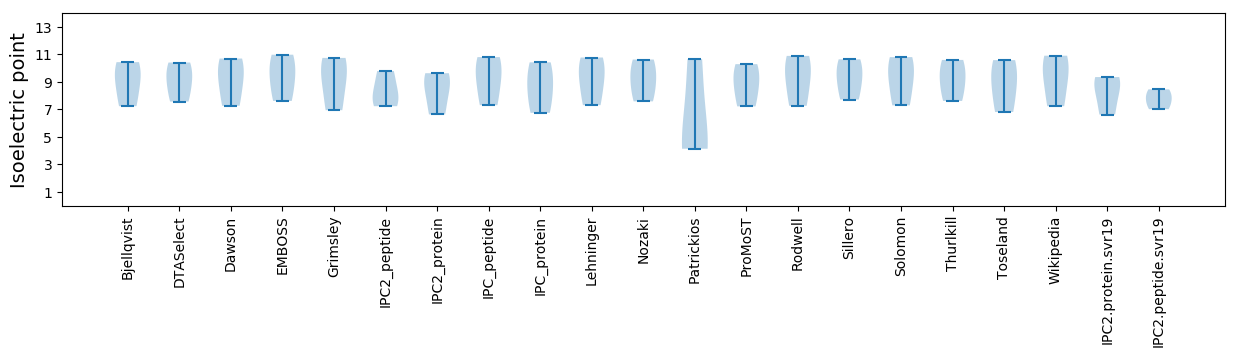
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A190WHA1|A0A190WHA1_9CIRC Uncharacterized protein OS=Circovirus-like genome DCCV-2 OX=1788442 PE=4 SV=1
MM1 pKa = 7.31FRR3 pKa = 11.84INAKK7 pKa = 9.98NFSLTYY13 pKa = 10.92SNVEE17 pKa = 4.04QQSGVAWLPWDD28 pKa = 3.64KK29 pKa = 11.2EE30 pKa = 4.07SLLRR34 pKa = 11.84HH35 pKa = 6.15LEE37 pKa = 3.98SLGDD41 pKa = 3.77GVIGMVSRR49 pKa = 11.84EE50 pKa = 3.85QHH52 pKa = 5.54QDD54 pKa = 2.74GSTHH58 pKa = 4.74FHH60 pKa = 6.47AWVQFPRR67 pKa = 11.84KK68 pKa = 9.25RR69 pKa = 11.84DD70 pKa = 3.08IRR72 pKa = 11.84ASNFFDD78 pKa = 3.37WQGCHH83 pKa = 6.65PNVQATNNLRR93 pKa = 11.84AWKK96 pKa = 8.97TYY98 pKa = 8.23ICKK101 pKa = 10.71DD102 pKa = 3.17GDD104 pKa = 4.02HH105 pKa = 6.51TPEE108 pKa = 4.73PATPTATGDD117 pKa = 3.94LFVLCHH123 pKa = 6.76SMEE126 pKa = 3.72KK127 pKa = 10.49HH128 pKa = 5.21EE129 pKa = 4.24WVNYY133 pKa = 10.26CIGKK137 pKa = 10.12KK138 pKa = 9.69IGFQYY143 pKa = 10.01MEE145 pKa = 5.02YY146 pKa = 9.72IWKK149 pKa = 9.29DD150 pKa = 3.17CHH152 pKa = 6.4MDD154 pKa = 3.43RR155 pKa = 11.84DD156 pKa = 4.15STIEE160 pKa = 4.15DD161 pKa = 3.79NPPPEE166 pKa = 5.11AVMCAALEE174 pKa = 4.0QFIYY178 pKa = 10.23PSLDD182 pKa = 3.04RR183 pKa = 11.84SPLVLVGDD191 pKa = 4.32TGCGKK196 pKa = 6.67TTWAIKK202 pKa = 9.8NAPKK206 pKa = 9.98PICFVSHH213 pKa = 6.58MDD215 pKa = 3.52QLKK218 pKa = 10.68HH219 pKa = 6.9FDD221 pKa = 3.74PQYY224 pKa = 10.34HH225 pKa = 6.25KK226 pKa = 11.08SIVFDD231 pKa = 4.6DD232 pKa = 4.02MDD234 pKa = 4.55FKK236 pKa = 11.14HH237 pKa = 6.65LPRR240 pKa = 11.84EE241 pKa = 4.28SQIHH245 pKa = 5.91ILDD248 pKa = 3.5MTTPRR253 pKa = 11.84AIHH256 pKa = 6.03RR257 pKa = 11.84RR258 pKa = 11.84YY259 pKa = 10.17GVTVIPAGTKK269 pKa = 10.18KK270 pKa = 10.36IFTANSNPFIEE281 pKa = 5.23DD282 pKa = 3.05PAIIRR287 pKa = 11.84RR288 pKa = 11.84RR289 pKa = 11.84TLRR292 pKa = 11.84IVQGLL297 pKa = 3.52
MM1 pKa = 7.31FRR3 pKa = 11.84INAKK7 pKa = 9.98NFSLTYY13 pKa = 10.92SNVEE17 pKa = 4.04QQSGVAWLPWDD28 pKa = 3.64KK29 pKa = 11.2EE30 pKa = 4.07SLLRR34 pKa = 11.84HH35 pKa = 6.15LEE37 pKa = 3.98SLGDD41 pKa = 3.77GVIGMVSRR49 pKa = 11.84EE50 pKa = 3.85QHH52 pKa = 5.54QDD54 pKa = 2.74GSTHH58 pKa = 4.74FHH60 pKa = 6.47AWVQFPRR67 pKa = 11.84KK68 pKa = 9.25RR69 pKa = 11.84DD70 pKa = 3.08IRR72 pKa = 11.84ASNFFDD78 pKa = 3.37WQGCHH83 pKa = 6.65PNVQATNNLRR93 pKa = 11.84AWKK96 pKa = 8.97TYY98 pKa = 8.23ICKK101 pKa = 10.71DD102 pKa = 3.17GDD104 pKa = 4.02HH105 pKa = 6.51TPEE108 pKa = 4.73PATPTATGDD117 pKa = 3.94LFVLCHH123 pKa = 6.76SMEE126 pKa = 3.72KK127 pKa = 10.49HH128 pKa = 5.21EE129 pKa = 4.24WVNYY133 pKa = 10.26CIGKK137 pKa = 10.12KK138 pKa = 9.69IGFQYY143 pKa = 10.01MEE145 pKa = 5.02YY146 pKa = 9.72IWKK149 pKa = 9.29DD150 pKa = 3.17CHH152 pKa = 6.4MDD154 pKa = 3.43RR155 pKa = 11.84DD156 pKa = 4.15STIEE160 pKa = 4.15DD161 pKa = 3.79NPPPEE166 pKa = 5.11AVMCAALEE174 pKa = 4.0QFIYY178 pKa = 10.23PSLDD182 pKa = 3.04RR183 pKa = 11.84SPLVLVGDD191 pKa = 4.32TGCGKK196 pKa = 6.67TTWAIKK202 pKa = 9.8NAPKK206 pKa = 9.98PICFVSHH213 pKa = 6.58MDD215 pKa = 3.52QLKK218 pKa = 10.68HH219 pKa = 6.9FDD221 pKa = 3.74PQYY224 pKa = 10.34HH225 pKa = 6.25KK226 pKa = 11.08SIVFDD231 pKa = 4.6DD232 pKa = 4.02MDD234 pKa = 4.55FKK236 pKa = 11.14HH237 pKa = 6.65LPRR240 pKa = 11.84EE241 pKa = 4.28SQIHH245 pKa = 5.91ILDD248 pKa = 3.5MTTPRR253 pKa = 11.84AIHH256 pKa = 6.03RR257 pKa = 11.84RR258 pKa = 11.84YY259 pKa = 10.17GVTVIPAGTKK269 pKa = 10.18KK270 pKa = 10.36IFTANSNPFIEE281 pKa = 5.23DD282 pKa = 3.05PAIIRR287 pKa = 11.84RR288 pKa = 11.84RR289 pKa = 11.84TLRR292 pKa = 11.84IVQGLL297 pKa = 3.52
Molecular weight: 34.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A190WHI4|A0A190WHI4_9CIRC Uncharacterized protein OS=Circovirus-like genome DCCV-2 OX=1788442 PE=4 SV=1
MM1 pKa = 7.46LKK3 pKa = 10.06ISALPIVMLNNNPGSLGYY21 pKa = 9.68PGIKK25 pKa = 9.24NHH27 pKa = 6.59CSVTLSPLEE36 pKa = 4.14TEE38 pKa = 4.22SLVWCLANNTRR49 pKa = 11.84MEE51 pKa = 4.35VPISTPGFNFLEE63 pKa = 4.36RR64 pKa = 11.84GIYY67 pKa = 9.13EE68 pKa = 4.63PPTSSTGRR76 pKa = 11.84GAIQTSRR83 pKa = 11.84LLITCGRR90 pKa = 11.84GKK92 pKa = 9.87PISAKK97 pKa = 9.07TATIRR102 pKa = 11.84RR103 pKa = 11.84NRR105 pKa = 11.84LRR107 pKa = 11.84RR108 pKa = 11.84LRR110 pKa = 11.84RR111 pKa = 11.84VTYY114 pKa = 9.33LCCATAWKK122 pKa = 8.19NTSGSTIAA130 pKa = 4.87
MM1 pKa = 7.46LKK3 pKa = 10.06ISALPIVMLNNNPGSLGYY21 pKa = 9.68PGIKK25 pKa = 9.24NHH27 pKa = 6.59CSVTLSPLEE36 pKa = 4.14TEE38 pKa = 4.22SLVWCLANNTRR49 pKa = 11.84MEE51 pKa = 4.35VPISTPGFNFLEE63 pKa = 4.36RR64 pKa = 11.84GIYY67 pKa = 9.13EE68 pKa = 4.63PPTSSTGRR76 pKa = 11.84GAIQTSRR83 pKa = 11.84LLITCGRR90 pKa = 11.84GKK92 pKa = 9.87PISAKK97 pKa = 9.07TATIRR102 pKa = 11.84RR103 pKa = 11.84NRR105 pKa = 11.84LRR107 pKa = 11.84RR108 pKa = 11.84LRR110 pKa = 11.84RR111 pKa = 11.84VTYY114 pKa = 9.33LCCATAWKK122 pKa = 8.19NTSGSTIAA130 pKa = 4.87
Molecular weight: 14.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
678 |
130 |
297 |
226.0 |
25.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.49 ± 0.514 | 2.212 ± 0.616 |
4.72 ± 1.402 | 3.097 ± 0.889 |
5.015 ± 0.891 | 5.752 ± 0.448 |
2.802 ± 1.23 | 5.605 ± 1.241 |
4.867 ± 0.468 | 7.08 ± 0.872 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.802 ± 0.14 | 5.9 ± 1.045 |
5.9 ± 0.487 | 4.277 ± 0.883 |
5.605 ± 0.975 | 6.785 ± 0.8 |
10.029 ± 2.35 | 6.047 ± 0.836 |
1.622 ± 0.646 | 3.392 ± 0.61 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
