
Wuhan Insect virus 5
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus; Wuhan 5 insect cytorhabdovirus
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
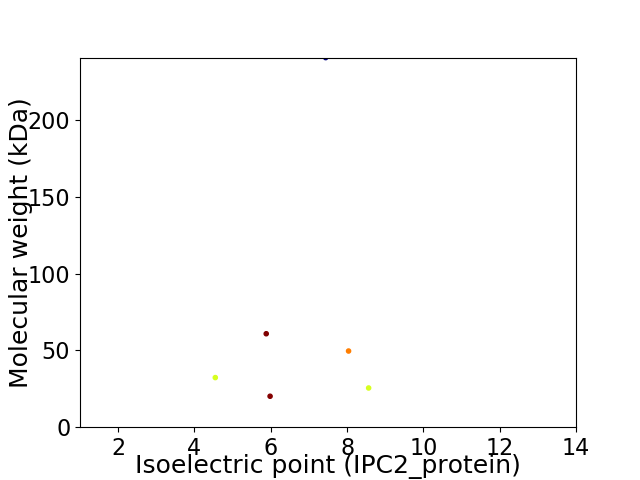
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0B5KTM4|A0A0B5KTM4_9RHAB 4b protein OS=Wuhan Insect virus 5 OX=1608110 GN=4b PE=4 SV=1
MM1 pKa = 7.26VDD3 pKa = 3.21KK4 pKa = 11.11VPNEE8 pKa = 3.59FAGAFNPDD16 pKa = 2.83GKK18 pKa = 9.37TDD20 pKa = 4.39FSSLADD26 pKa = 3.79NLDD29 pKa = 3.87DD30 pKa = 5.1SVDD33 pKa = 4.13NIPDD37 pKa = 3.96PSLSIPTQARR47 pKa = 11.84EE48 pKa = 4.45AVDD51 pKa = 3.64QPRR54 pKa = 11.84KK55 pKa = 9.95QPVGDD60 pKa = 3.66VAATLLCLKK69 pKa = 9.02NTAHH73 pKa = 6.0NHH75 pKa = 4.55GAIVTNDD82 pKa = 3.15MEE84 pKa = 5.28SLFKK88 pKa = 10.57YY89 pKa = 10.24YY90 pKa = 10.35CANEE94 pKa = 3.84QLDD97 pKa = 3.42IRR99 pKa = 11.84DD100 pKa = 2.91IEE102 pKa = 4.43FFVRR106 pKa = 11.84GYY108 pKa = 11.11VYY110 pKa = 10.25ATGSQVGPKK119 pKa = 9.4IQEE122 pKa = 3.76ATEE125 pKa = 4.05GLKK128 pKa = 10.45VEE130 pKa = 4.05MRR132 pKa = 11.84AIQRR136 pKa = 11.84ANATLAEE143 pKa = 4.45TIKK146 pKa = 11.04LLANQAVSVEE156 pKa = 4.15KK157 pKa = 10.47EE158 pKa = 3.53IAALSVNVKK167 pKa = 10.58NDD169 pKa = 2.8VTAALKK175 pKa = 10.79SAVAGKK181 pKa = 10.37AKK183 pKa = 10.67LGDD186 pKa = 3.77QPVSPLQTVKK196 pKa = 10.98LPLAKK201 pKa = 10.49ASLEE205 pKa = 4.2VVAAPNPVPATLEE218 pKa = 3.76VGEE221 pKa = 4.23PSKK224 pKa = 10.83PSEE227 pKa = 4.46TGLSDD232 pKa = 3.92PSQLKK237 pKa = 10.28KK238 pKa = 9.82MRR240 pKa = 11.84SVLSMIGVEE249 pKa = 4.73DD250 pKa = 3.56AVLNNLLDD258 pKa = 3.61NDD260 pKa = 3.84IPVIYY265 pKa = 9.02PAAIMEE271 pKa = 4.53EE272 pKa = 4.29YY273 pKa = 11.05GNLLGDD279 pKa = 4.44PEE281 pKa = 4.45VCDD284 pKa = 4.55LIKK287 pKa = 10.9EE288 pKa = 4.18EE289 pKa = 4.19VMGNIEE295 pKa = 4.95KK296 pKa = 10.59FLLCGAA302 pKa = 4.91
MM1 pKa = 7.26VDD3 pKa = 3.21KK4 pKa = 11.11VPNEE8 pKa = 3.59FAGAFNPDD16 pKa = 2.83GKK18 pKa = 9.37TDD20 pKa = 4.39FSSLADD26 pKa = 3.79NLDD29 pKa = 3.87DD30 pKa = 5.1SVDD33 pKa = 4.13NIPDD37 pKa = 3.96PSLSIPTQARR47 pKa = 11.84EE48 pKa = 4.45AVDD51 pKa = 3.64QPRR54 pKa = 11.84KK55 pKa = 9.95QPVGDD60 pKa = 3.66VAATLLCLKK69 pKa = 9.02NTAHH73 pKa = 6.0NHH75 pKa = 4.55GAIVTNDD82 pKa = 3.15MEE84 pKa = 5.28SLFKK88 pKa = 10.57YY89 pKa = 10.24YY90 pKa = 10.35CANEE94 pKa = 3.84QLDD97 pKa = 3.42IRR99 pKa = 11.84DD100 pKa = 2.91IEE102 pKa = 4.43FFVRR106 pKa = 11.84GYY108 pKa = 11.11VYY110 pKa = 10.25ATGSQVGPKK119 pKa = 9.4IQEE122 pKa = 3.76ATEE125 pKa = 4.05GLKK128 pKa = 10.45VEE130 pKa = 4.05MRR132 pKa = 11.84AIQRR136 pKa = 11.84ANATLAEE143 pKa = 4.45TIKK146 pKa = 11.04LLANQAVSVEE156 pKa = 4.15KK157 pKa = 10.47EE158 pKa = 3.53IAALSVNVKK167 pKa = 10.58NDD169 pKa = 2.8VTAALKK175 pKa = 10.79SAVAGKK181 pKa = 10.37AKK183 pKa = 10.67LGDD186 pKa = 3.77QPVSPLQTVKK196 pKa = 10.98LPLAKK201 pKa = 10.49ASLEE205 pKa = 4.2VVAAPNPVPATLEE218 pKa = 3.76VGEE221 pKa = 4.23PSKK224 pKa = 10.83PSEE227 pKa = 4.46TGLSDD232 pKa = 3.92PSQLKK237 pKa = 10.28KK238 pKa = 9.82MRR240 pKa = 11.84SVLSMIGVEE249 pKa = 4.73DD250 pKa = 3.56AVLNNLLDD258 pKa = 3.61NDD260 pKa = 3.84IPVIYY265 pKa = 9.02PAAIMEE271 pKa = 4.53EE272 pKa = 4.29YY273 pKa = 11.05GNLLGDD279 pKa = 4.44PEE281 pKa = 4.45VCDD284 pKa = 4.55LIKK287 pKa = 10.9EE288 pKa = 4.18EE289 pKa = 4.19VMGNIEE295 pKa = 4.95KK296 pKa = 10.59FLLCGAA302 pKa = 4.91
Molecular weight: 32.3 kDa
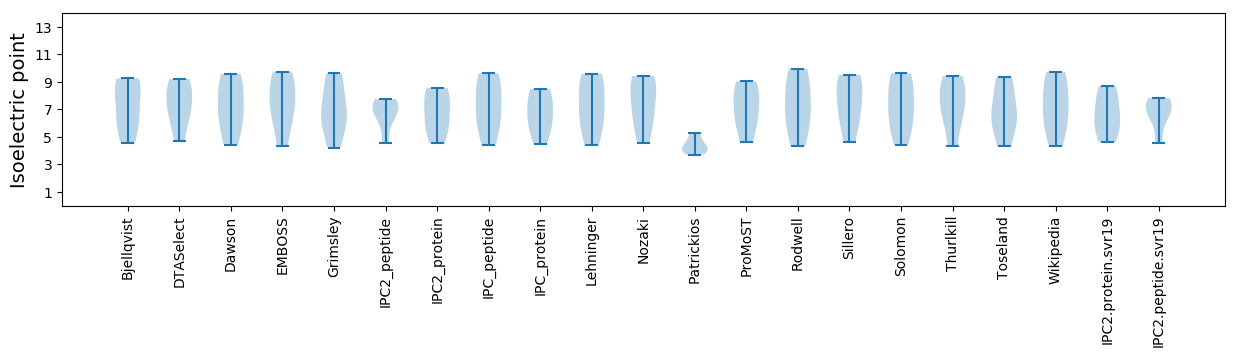
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KXN2|A0A0B5KXN2_9RHAB Matrix protein OS=Wuhan Insect virus 5 OX=1608110 GN=M PE=4 SV=1
MM1 pKa = 7.59EE2 pKa = 4.47SQMVVSSGEE11 pKa = 3.91FTMGEE16 pKa = 3.93TDD18 pKa = 4.29GVISLSKK25 pKa = 10.64KK26 pKa = 9.63IGLFQKK32 pKa = 10.81LLTKK36 pKa = 10.8FGYY39 pKa = 9.39TNIAVHH45 pKa = 6.12QVIFKK50 pKa = 10.43YY51 pKa = 10.37KK52 pKa = 9.3SRR54 pKa = 11.84CPAGAEE60 pKa = 3.96GLMTLKK66 pKa = 10.26MVDD69 pKa = 3.81RR70 pKa = 11.84RR71 pKa = 11.84LDD73 pKa = 3.89DD74 pKa = 5.12PDD76 pKa = 6.34DD77 pKa = 3.83MMIDD81 pKa = 3.55GLSFDD86 pKa = 3.76VKK88 pKa = 10.91DD89 pKa = 3.66WIVFSWSYY97 pKa = 10.54PCWFHH102 pKa = 7.66YY103 pKa = 10.69KK104 pKa = 10.54DD105 pKa = 4.13FEE107 pKa = 4.67SKK109 pKa = 10.73EE110 pKa = 3.95KK111 pKa = 10.8DD112 pKa = 3.33LLEE115 pKa = 4.49IEE117 pKa = 4.37WGVSGSNMIDD127 pKa = 3.51SVSLGHH133 pKa = 5.96YY134 pKa = 8.69KK135 pKa = 10.57VKK137 pKa = 10.2IAYY140 pKa = 9.5KK141 pKa = 8.9MRR143 pKa = 11.84NDD145 pKa = 2.89ITRR148 pKa = 11.84LVSNPNIAQKK158 pKa = 10.29VHH160 pKa = 5.8SQVHH164 pKa = 4.04VSKK167 pKa = 10.5FVKK170 pKa = 9.7KK171 pKa = 10.35QKK173 pKa = 9.26TGRR176 pKa = 11.84LSKK179 pKa = 10.19STGNLLNVSSPPDD192 pKa = 3.27SGMKK196 pKa = 8.08NTKK199 pKa = 9.95NSSDD203 pKa = 3.53YY204 pKa = 10.88RR205 pKa = 11.84KK206 pKa = 10.45EE207 pKa = 3.87EE208 pKa = 4.1LLTVSAEE215 pKa = 3.82RR216 pKa = 11.84LFPIRR221 pKa = 11.84GVKK224 pKa = 8.97PP225 pKa = 3.34
MM1 pKa = 7.59EE2 pKa = 4.47SQMVVSSGEE11 pKa = 3.91FTMGEE16 pKa = 3.93TDD18 pKa = 4.29GVISLSKK25 pKa = 10.64KK26 pKa = 9.63IGLFQKK32 pKa = 10.81LLTKK36 pKa = 10.8FGYY39 pKa = 9.39TNIAVHH45 pKa = 6.12QVIFKK50 pKa = 10.43YY51 pKa = 10.37KK52 pKa = 9.3SRR54 pKa = 11.84CPAGAEE60 pKa = 3.96GLMTLKK66 pKa = 10.26MVDD69 pKa = 3.81RR70 pKa = 11.84RR71 pKa = 11.84LDD73 pKa = 3.89DD74 pKa = 5.12PDD76 pKa = 6.34DD77 pKa = 3.83MMIDD81 pKa = 3.55GLSFDD86 pKa = 3.76VKK88 pKa = 10.91DD89 pKa = 3.66WIVFSWSYY97 pKa = 10.54PCWFHH102 pKa = 7.66YY103 pKa = 10.69KK104 pKa = 10.54DD105 pKa = 4.13FEE107 pKa = 4.67SKK109 pKa = 10.73EE110 pKa = 3.95KK111 pKa = 10.8DD112 pKa = 3.33LLEE115 pKa = 4.49IEE117 pKa = 4.37WGVSGSNMIDD127 pKa = 3.51SVSLGHH133 pKa = 5.96YY134 pKa = 8.69KK135 pKa = 10.57VKK137 pKa = 10.2IAYY140 pKa = 9.5KK141 pKa = 8.9MRR143 pKa = 11.84NDD145 pKa = 2.89ITRR148 pKa = 11.84LVSNPNIAQKK158 pKa = 10.29VHH160 pKa = 5.8SQVHH164 pKa = 4.04VSKK167 pKa = 10.5FVKK170 pKa = 9.7KK171 pKa = 10.35QKK173 pKa = 9.26TGRR176 pKa = 11.84LSKK179 pKa = 10.19STGNLLNVSSPPDD192 pKa = 3.27SGMKK196 pKa = 8.08NTKK199 pKa = 9.95NSSDD203 pKa = 3.53YY204 pKa = 10.88RR205 pKa = 11.84KK206 pKa = 10.45EE207 pKa = 3.87EE208 pKa = 4.1LLTVSAEE215 pKa = 3.82RR216 pKa = 11.84LFPIRR221 pKa = 11.84GVKK224 pKa = 8.97PP225 pKa = 3.34
Molecular weight: 25.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3814 |
187 |
2098 |
635.7 |
71.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.821 ± 1.454 | 2.071 ± 0.352 |
5.952 ± 0.211 | 5.69 ± 0.219 |
3.644 ± 0.279 | 5.821 ± 0.346 |
2.045 ± 0.29 | 6.319 ± 0.517 |
6.266 ± 0.782 | 9.255 ± 0.606 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.644 ± 0.402 | 3.723 ± 0.325 |
4.51 ± 0.462 | 2.989 ± 0.646 |
5.48 ± 1.165 | 9.491 ± 0.469 |
5.506 ± 0.31 | 6.738 ± 0.5 |
1.363 ± 0.261 | 3.671 ± 0.476 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
