
Hubei tombus-like virus 16
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.48
Get precalculated fractions of proteins
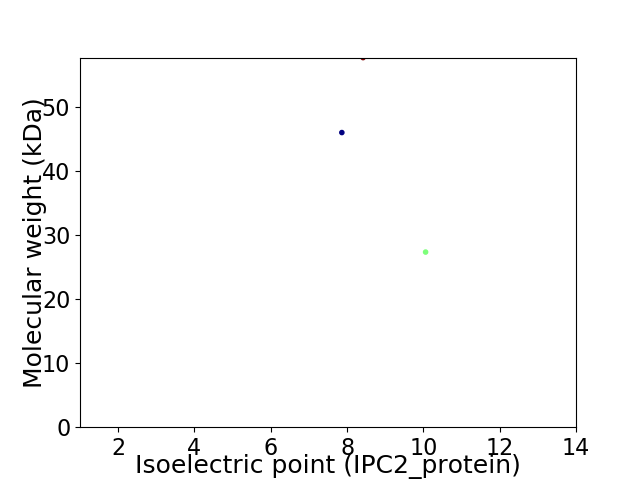
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KG84|A0A1L3KG84_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 16 OX=1923262 PE=4 SV=1
MM1 pKa = 7.15FVKK4 pKa = 10.04PDD6 pKa = 3.75KK7 pKa = 10.94IEE9 pKa = 4.18AGEE12 pKa = 4.23IQDD15 pKa = 3.69KK16 pKa = 10.59APRR19 pKa = 11.84AIQYY23 pKa = 8.9RR24 pKa = 11.84GPAYY28 pKa = 9.82NLEE31 pKa = 4.07MLRR34 pKa = 11.84YY35 pKa = 9.34IKK37 pKa = 10.46PFEE40 pKa = 4.09HH41 pKa = 7.1KK42 pKa = 10.29IYY44 pKa = 10.92DD45 pKa = 3.83EE46 pKa = 4.21LHH48 pKa = 6.16LGNYY52 pKa = 9.1SRR54 pKa = 11.84TRR56 pKa = 11.84AIVKK60 pKa = 9.61GLNNYY65 pKa = 7.99QRR67 pKa = 11.84AEE69 pKa = 3.95LFFEE73 pKa = 4.38KK74 pKa = 10.31LQNFQQPAFLLIDD87 pKa = 3.46HH88 pKa = 6.92SKK90 pKa = 10.17FDD92 pKa = 3.34STVRR96 pKa = 11.84VEE98 pKa = 4.47HH99 pKa = 6.44LRR101 pKa = 11.84STHH104 pKa = 5.37RR105 pKa = 11.84KK106 pKa = 7.89YY107 pKa = 10.57MKK109 pKa = 10.48AFNSKK114 pKa = 10.13HH115 pKa = 5.62LWQLCKK121 pKa = 10.74VQISNKK127 pKa = 9.33CYY129 pKa = 9.57SKK131 pKa = 11.25NGIHH135 pKa = 5.03YY136 pKa = 6.93TSRR139 pKa = 11.84GTRR142 pKa = 11.84MSGDD146 pKa = 3.84PDD148 pKa = 3.89TGCGNSVINGDD159 pKa = 4.12CLHH162 pKa = 6.46EE163 pKa = 4.17VLRR166 pKa = 11.84VSGIAKK172 pKa = 9.95YY173 pKa = 10.62EE174 pKa = 3.83IMLDD178 pKa = 3.65GDD180 pKa = 3.89DD181 pKa = 4.67SIVIIEE187 pKa = 4.53KK188 pKa = 10.35GALSAFKK195 pKa = 10.22YY196 pKa = 10.72DD197 pKa = 2.96HH198 pKa = 6.76FEE200 pKa = 3.88RR201 pKa = 11.84LGFEE205 pKa = 4.18TKK207 pKa = 9.72MSVVYY212 pKa = 10.05DD213 pKa = 3.34YY214 pKa = 11.68HH215 pKa = 7.71EE216 pKa = 4.48VEE218 pKa = 4.83FCRR221 pKa = 11.84SKK223 pKa = 11.33PMMHH227 pKa = 7.07PRR229 pKa = 11.84PCFIRR234 pKa = 11.84DD235 pKa = 3.62PKK237 pKa = 10.12RR238 pKa = 11.84TISNTMMCLKK248 pKa = 10.38HH249 pKa = 5.72YY250 pKa = 10.83AEE252 pKa = 5.16RR253 pKa = 11.84DD254 pKa = 3.74YY255 pKa = 11.43KK256 pKa = 10.97SWLAAVGLCEE266 pKa = 4.33LACNDD271 pKa = 3.87GVPVISVLGRR281 pKa = 11.84TLSGFSKK288 pKa = 10.55RR289 pKa = 11.84KK290 pKa = 10.12LFDD293 pKa = 4.38EE294 pKa = 4.3DD295 pKa = 4.48TLWKK299 pKa = 9.54MGNLAGGKK307 pKa = 9.47YY308 pKa = 7.49DD309 pKa = 3.7TPITTEE315 pKa = 3.54SRR317 pKa = 11.84LEE319 pKa = 3.86YY320 pKa = 11.1AMTWGVDD327 pKa = 3.62CEE329 pKa = 4.36VQLLLEE335 pKa = 4.59EE336 pKa = 4.26EE337 pKa = 4.71FKK339 pKa = 10.97TSIDD343 pKa = 4.81DD344 pKa = 3.56SFHH347 pKa = 5.0YY348 pKa = 9.98CKK350 pKa = 10.58RR351 pKa = 11.84IPRR354 pKa = 11.84VKK356 pKa = 10.14QRR358 pKa = 11.84TNIKK362 pKa = 8.16STRR365 pKa = 11.84EE366 pKa = 3.84NFNGRR371 pKa = 11.84TTVSVWTAASAYY383 pKa = 9.99QSLDD387 pKa = 3.36QFSSSSWWCSGG398 pKa = 3.02
MM1 pKa = 7.15FVKK4 pKa = 10.04PDD6 pKa = 3.75KK7 pKa = 10.94IEE9 pKa = 4.18AGEE12 pKa = 4.23IQDD15 pKa = 3.69KK16 pKa = 10.59APRR19 pKa = 11.84AIQYY23 pKa = 8.9RR24 pKa = 11.84GPAYY28 pKa = 9.82NLEE31 pKa = 4.07MLRR34 pKa = 11.84YY35 pKa = 9.34IKK37 pKa = 10.46PFEE40 pKa = 4.09HH41 pKa = 7.1KK42 pKa = 10.29IYY44 pKa = 10.92DD45 pKa = 3.83EE46 pKa = 4.21LHH48 pKa = 6.16LGNYY52 pKa = 9.1SRR54 pKa = 11.84TRR56 pKa = 11.84AIVKK60 pKa = 9.61GLNNYY65 pKa = 7.99QRR67 pKa = 11.84AEE69 pKa = 3.95LFFEE73 pKa = 4.38KK74 pKa = 10.31LQNFQQPAFLLIDD87 pKa = 3.46HH88 pKa = 6.92SKK90 pKa = 10.17FDD92 pKa = 3.34STVRR96 pKa = 11.84VEE98 pKa = 4.47HH99 pKa = 6.44LRR101 pKa = 11.84STHH104 pKa = 5.37RR105 pKa = 11.84KK106 pKa = 7.89YY107 pKa = 10.57MKK109 pKa = 10.48AFNSKK114 pKa = 10.13HH115 pKa = 5.62LWQLCKK121 pKa = 10.74VQISNKK127 pKa = 9.33CYY129 pKa = 9.57SKK131 pKa = 11.25NGIHH135 pKa = 5.03YY136 pKa = 6.93TSRR139 pKa = 11.84GTRR142 pKa = 11.84MSGDD146 pKa = 3.84PDD148 pKa = 3.89TGCGNSVINGDD159 pKa = 4.12CLHH162 pKa = 6.46EE163 pKa = 4.17VLRR166 pKa = 11.84VSGIAKK172 pKa = 9.95YY173 pKa = 10.62EE174 pKa = 3.83IMLDD178 pKa = 3.65GDD180 pKa = 3.89DD181 pKa = 4.67SIVIIEE187 pKa = 4.53KK188 pKa = 10.35GALSAFKK195 pKa = 10.22YY196 pKa = 10.72DD197 pKa = 2.96HH198 pKa = 6.76FEE200 pKa = 3.88RR201 pKa = 11.84LGFEE205 pKa = 4.18TKK207 pKa = 9.72MSVVYY212 pKa = 10.05DD213 pKa = 3.34YY214 pKa = 11.68HH215 pKa = 7.71EE216 pKa = 4.48VEE218 pKa = 4.83FCRR221 pKa = 11.84SKK223 pKa = 11.33PMMHH227 pKa = 7.07PRR229 pKa = 11.84PCFIRR234 pKa = 11.84DD235 pKa = 3.62PKK237 pKa = 10.12RR238 pKa = 11.84TISNTMMCLKK248 pKa = 10.38HH249 pKa = 5.72YY250 pKa = 10.83AEE252 pKa = 5.16RR253 pKa = 11.84DD254 pKa = 3.74YY255 pKa = 11.43KK256 pKa = 10.97SWLAAVGLCEE266 pKa = 4.33LACNDD271 pKa = 3.87GVPVISVLGRR281 pKa = 11.84TLSGFSKK288 pKa = 10.55RR289 pKa = 11.84KK290 pKa = 10.12LFDD293 pKa = 4.38EE294 pKa = 4.3DD295 pKa = 4.48TLWKK299 pKa = 9.54MGNLAGGKK307 pKa = 9.47YY308 pKa = 7.49DD309 pKa = 3.7TPITTEE315 pKa = 3.54SRR317 pKa = 11.84LEE319 pKa = 3.86YY320 pKa = 11.1AMTWGVDD327 pKa = 3.62CEE329 pKa = 4.36VQLLLEE335 pKa = 4.59EE336 pKa = 4.26EE337 pKa = 4.71FKK339 pKa = 10.97TSIDD343 pKa = 4.81DD344 pKa = 3.56SFHH347 pKa = 5.0YY348 pKa = 9.98CKK350 pKa = 10.58RR351 pKa = 11.84IPRR354 pKa = 11.84VKK356 pKa = 10.14QRR358 pKa = 11.84TNIKK362 pKa = 8.16STRR365 pKa = 11.84EE366 pKa = 3.84NFNGRR371 pKa = 11.84TTVSVWTAASAYY383 pKa = 9.99QSLDD387 pKa = 3.36QFSSSSWWCSGG398 pKa = 3.02
Molecular weight: 46.0 kDa
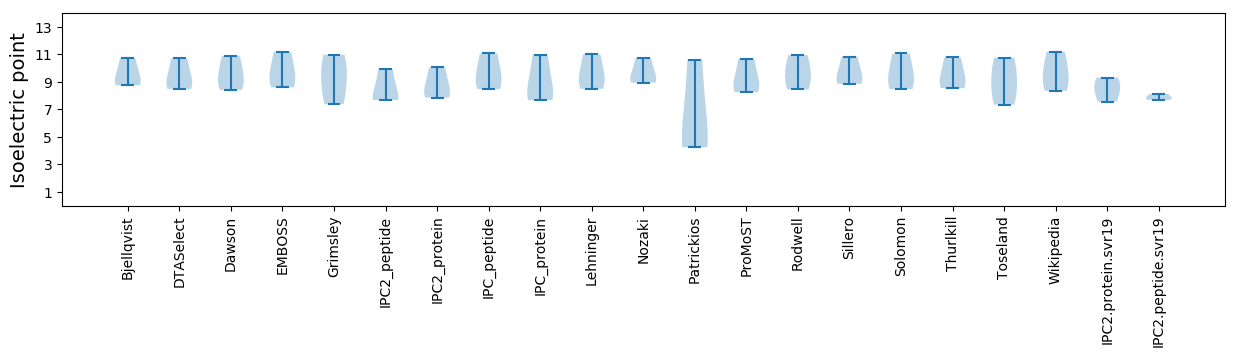
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGL2|A0A1L3KGL2_9VIRU Capsid protein OS=Hubei tombus-like virus 16 OX=1923262 PE=3 SV=1
MM1 pKa = 7.79AGQLYY6 pKa = 9.33PYY8 pKa = 8.31GQPRR12 pKa = 11.84PRR14 pKa = 11.84INPWINLAAAAGGAAARR31 pKa = 11.84QLVNYY36 pKa = 8.11GANQFNAYY44 pKa = 8.5MRR46 pKa = 11.84GPPPPPRR53 pKa = 11.84GGRR56 pKa = 11.84RR57 pKa = 11.84GRR59 pKa = 11.84GRR61 pKa = 11.84RR62 pKa = 11.84SFNMQPMLAALPPPPPRR79 pKa = 11.84ARR81 pKa = 11.84RR82 pKa = 11.84GGRR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84TARR91 pKa = 11.84GSNAGGSPTQIFRR104 pKa = 11.84GTEE107 pKa = 3.77YY108 pKa = 10.66LAEE111 pKa = 4.36PTTTLATLQFNCASTTHH128 pKa = 6.42PRR130 pKa = 11.84LSNTAKK136 pKa = 9.96CYY138 pKa = 8.53EE139 pKa = 4.24RR140 pKa = 11.84YY141 pKa = 9.42RR142 pKa = 11.84VRR144 pKa = 11.84SVVLTYY150 pKa = 11.19NSTSGTATTGGVTLGIHH167 pKa = 6.74PGPTNSNVKK176 pKa = 10.02KK177 pKa = 10.88AEE179 pKa = 5.12DD180 pKa = 3.54ILKK183 pKa = 9.98LDD185 pKa = 4.17PSRR188 pKa = 11.84TTPAWKK194 pKa = 10.37NSTIRR199 pKa = 11.84LGQNIDD205 pKa = 3.13AQRR208 pKa = 11.84WMHH211 pKa = 5.75VGKK214 pKa = 7.67TTEE217 pKa = 3.9EE218 pKa = 4.4GVAFTCYY225 pKa = 10.09YY226 pKa = 9.76IKK228 pKa = 11.0SGDD231 pKa = 3.29ATGQIKK237 pKa = 8.25ITYY240 pKa = 8.9DD241 pKa = 2.83IEE243 pKa = 3.91FAYY246 pKa = 9.28PVPFF250 pKa = 4.67
MM1 pKa = 7.79AGQLYY6 pKa = 9.33PYY8 pKa = 8.31GQPRR12 pKa = 11.84PRR14 pKa = 11.84INPWINLAAAAGGAAARR31 pKa = 11.84QLVNYY36 pKa = 8.11GANQFNAYY44 pKa = 8.5MRR46 pKa = 11.84GPPPPPRR53 pKa = 11.84GGRR56 pKa = 11.84RR57 pKa = 11.84GRR59 pKa = 11.84GRR61 pKa = 11.84RR62 pKa = 11.84SFNMQPMLAALPPPPPRR79 pKa = 11.84ARR81 pKa = 11.84RR82 pKa = 11.84GGRR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84TARR91 pKa = 11.84GSNAGGSPTQIFRR104 pKa = 11.84GTEE107 pKa = 3.77YY108 pKa = 10.66LAEE111 pKa = 4.36PTTTLATLQFNCASTTHH128 pKa = 6.42PRR130 pKa = 11.84LSNTAKK136 pKa = 9.96CYY138 pKa = 8.53EE139 pKa = 4.24RR140 pKa = 11.84YY141 pKa = 9.42RR142 pKa = 11.84VRR144 pKa = 11.84SVVLTYY150 pKa = 11.19NSTSGTATTGGVTLGIHH167 pKa = 6.74PGPTNSNVKK176 pKa = 10.02KK177 pKa = 10.88AEE179 pKa = 5.12DD180 pKa = 3.54ILKK183 pKa = 9.98LDD185 pKa = 4.17PSRR188 pKa = 11.84TTPAWKK194 pKa = 10.37NSTIRR199 pKa = 11.84LGQNIDD205 pKa = 3.13AQRR208 pKa = 11.84WMHH211 pKa = 5.75VGKK214 pKa = 7.67TTEE217 pKa = 3.9EE218 pKa = 4.4GVAFTCYY225 pKa = 10.09YY226 pKa = 9.76IKK228 pKa = 11.0SGDD231 pKa = 3.29ATGQIKK237 pKa = 8.25ITYY240 pKa = 8.9DD241 pKa = 2.83IEE243 pKa = 3.91FAYY246 pKa = 9.28PVPFF250 pKa = 4.67
Molecular weight: 27.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1146 |
250 |
498 |
382.0 |
43.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.981 ± 1.116 | 1.832 ± 0.525 |
4.538 ± 0.788 | 6.457 ± 1.202 |
4.101 ± 0.41 | 5.934 ± 1.443 |
2.443 ± 0.433 | 4.974 ± 0.256 |
6.457 ± 0.954 | 7.941 ± 0.779 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.269 ± 0.336 | 4.45 ± 0.34 |
7.853 ± 2.06 | 3.578 ± 0.25 |
8.115 ± 0.815 | 5.41 ± 1.209 |
5.846 ± 1.237 | 5.236 ± 0.513 |
1.571 ± 0.116 | 4.014 ± 0.523 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
