
Operophtera brumata (winter moth)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Amphiesmenoptera; Lepidoptera; Glossata; Neolepidoptera; Heteroneura; Ditrysia;
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
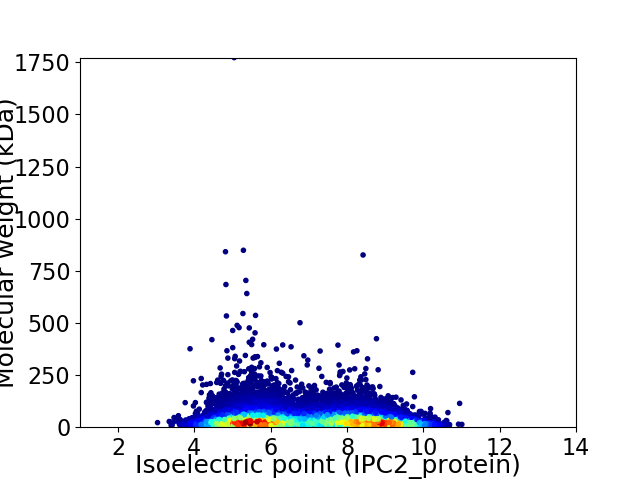
Virtual 2D-PAGE plot for 16814 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0L7LJQ0|A0A0L7LJQ0_9NEOP Putative alcohol dehydrogenase (Fragment) OS=Operophtera brumata OX=104452 GN=OBRU01_07362 PE=3 SV=1
MM1 pKa = 7.46IRR3 pKa = 11.84AQTQSSAVLSTVHH16 pKa = 6.48TDD18 pKa = 2.99SVYY21 pKa = 10.73PEE23 pKa = 4.44PYY25 pKa = 10.65VNNTWVQIEE34 pKa = 4.29ASVLMVTVRR43 pKa = 11.84QLEE46 pKa = 4.35SKK48 pKa = 9.33YY49 pKa = 9.54QQPQEE54 pKa = 4.46CPGIDD59 pKa = 3.29NSGSGNPRR67 pKa = 11.84EE68 pKa = 4.17TSTTDD73 pKa = 2.83EE74 pKa = 4.83RR75 pKa = 11.84LTEE78 pKa = 4.88DD79 pKa = 5.76DD80 pKa = 5.69DD81 pKa = 7.38DD82 pKa = 6.1EE83 pKa = 8.08DD84 pKa = 6.84DD85 pKa = 6.58DD86 pKa = 7.49DD87 pKa = 7.58DD88 pKa = 7.66DD89 pKa = 7.66DD90 pKa = 7.69DD91 pKa = 7.57DD92 pKa = 7.29DD93 pKa = 7.38DD94 pKa = 6.39EE95 pKa = 8.1DD96 pKa = 6.84DD97 pKa = 6.52DD98 pKa = 7.49DD99 pKa = 7.58DD100 pKa = 7.62DD101 pKa = 7.68DD102 pKa = 7.67DD103 pKa = 7.3DD104 pKa = 7.52DD105 pKa = 6.36DD106 pKa = 5.54VADD109 pKa = 6.12DD110 pKa = 6.2DD111 pKa = 6.65DD112 pKa = 7.74DD113 pKa = 7.66DD114 pKa = 7.65DD115 pKa = 7.66DD116 pKa = 7.65DD117 pKa = 7.65DD118 pKa = 7.61DD119 pKa = 7.63DD120 pKa = 7.45DD121 pKa = 7.47DD122 pKa = 6.6DD123 pKa = 6.67DD124 pKa = 4.21STVTINSMKK133 pKa = 10.48LNRR136 pKa = 11.84NNAIYY141 pKa = 9.38MGKK144 pKa = 9.52LLLMDD149 pKa = 4.18TPTTVFLIWVLYY161 pKa = 9.97IVFVLKK167 pKa = 10.88LGPQLMKK174 pKa = 10.77NRR176 pKa = 11.84PPYY179 pKa = 10.21NLNNILTIYY188 pKa = 10.43NALQVVSSAYY198 pKa = 9.21VVYY201 pKa = 10.07MGAPILWNYY210 pKa = 8.08GLKK213 pKa = 10.21PKK215 pKa = 10.37CLNNTDD221 pKa = 4.5DD222 pKa = 4.65KK223 pKa = 11.19IGYY226 pKa = 9.14KK227 pKa = 10.24FNQVTFLHH235 pKa = 6.63VYY237 pKa = 7.39HH238 pKa = 6.87HH239 pKa = 7.14SYY241 pKa = 11.13VVISSWFIIKK251 pKa = 10.44NDD253 pKa = 3.27TTYY256 pKa = 12.0AMLFPAVLNSFIHH269 pKa = 5.63VVMYY273 pKa = 8.8TYY275 pKa = 10.99YY276 pKa = 10.99GLSAFPALAKK286 pKa = 9.81YY287 pKa = 9.32LWWKK291 pKa = 10.54KK292 pKa = 10.61YY293 pKa = 7.51ITAMQMKK300 pKa = 9.59II301 pKa = 3.33
MM1 pKa = 7.46IRR3 pKa = 11.84AQTQSSAVLSTVHH16 pKa = 6.48TDD18 pKa = 2.99SVYY21 pKa = 10.73PEE23 pKa = 4.44PYY25 pKa = 10.65VNNTWVQIEE34 pKa = 4.29ASVLMVTVRR43 pKa = 11.84QLEE46 pKa = 4.35SKK48 pKa = 9.33YY49 pKa = 9.54QQPQEE54 pKa = 4.46CPGIDD59 pKa = 3.29NSGSGNPRR67 pKa = 11.84EE68 pKa = 4.17TSTTDD73 pKa = 2.83EE74 pKa = 4.83RR75 pKa = 11.84LTEE78 pKa = 4.88DD79 pKa = 5.76DD80 pKa = 5.69DD81 pKa = 7.38DD82 pKa = 6.1EE83 pKa = 8.08DD84 pKa = 6.84DD85 pKa = 6.58DD86 pKa = 7.49DD87 pKa = 7.58DD88 pKa = 7.66DD89 pKa = 7.66DD90 pKa = 7.69DD91 pKa = 7.57DD92 pKa = 7.29DD93 pKa = 7.38DD94 pKa = 6.39EE95 pKa = 8.1DD96 pKa = 6.84DD97 pKa = 6.52DD98 pKa = 7.49DD99 pKa = 7.58DD100 pKa = 7.62DD101 pKa = 7.68DD102 pKa = 7.67DD103 pKa = 7.3DD104 pKa = 7.52DD105 pKa = 6.36DD106 pKa = 5.54VADD109 pKa = 6.12DD110 pKa = 6.2DD111 pKa = 6.65DD112 pKa = 7.74DD113 pKa = 7.66DD114 pKa = 7.65DD115 pKa = 7.66DD116 pKa = 7.65DD117 pKa = 7.65DD118 pKa = 7.61DD119 pKa = 7.63DD120 pKa = 7.45DD121 pKa = 7.47DD122 pKa = 6.6DD123 pKa = 6.67DD124 pKa = 4.21STVTINSMKK133 pKa = 10.48LNRR136 pKa = 11.84NNAIYY141 pKa = 9.38MGKK144 pKa = 9.52LLLMDD149 pKa = 4.18TPTTVFLIWVLYY161 pKa = 9.97IVFVLKK167 pKa = 10.88LGPQLMKK174 pKa = 10.77NRR176 pKa = 11.84PPYY179 pKa = 10.21NLNNILTIYY188 pKa = 10.43NALQVVSSAYY198 pKa = 9.21VVYY201 pKa = 10.07MGAPILWNYY210 pKa = 8.08GLKK213 pKa = 10.21PKK215 pKa = 10.37CLNNTDD221 pKa = 4.5DD222 pKa = 4.65KK223 pKa = 11.19IGYY226 pKa = 9.14KK227 pKa = 10.24FNQVTFLHH235 pKa = 6.63VYY237 pKa = 7.39HH238 pKa = 6.87HH239 pKa = 7.14SYY241 pKa = 11.13VVISSWFIIKK251 pKa = 10.44NDD253 pKa = 3.27TTYY256 pKa = 12.0AMLFPAVLNSFIHH269 pKa = 5.63VVMYY273 pKa = 8.8TYY275 pKa = 10.99YY276 pKa = 10.99GLSAFPALAKK286 pKa = 9.81YY287 pKa = 9.32LWWKK291 pKa = 10.54KK292 pKa = 10.61YY293 pKa = 7.51ITAMQMKK300 pKa = 9.59II301 pKa = 3.33
Molecular weight: 34.55 kDa
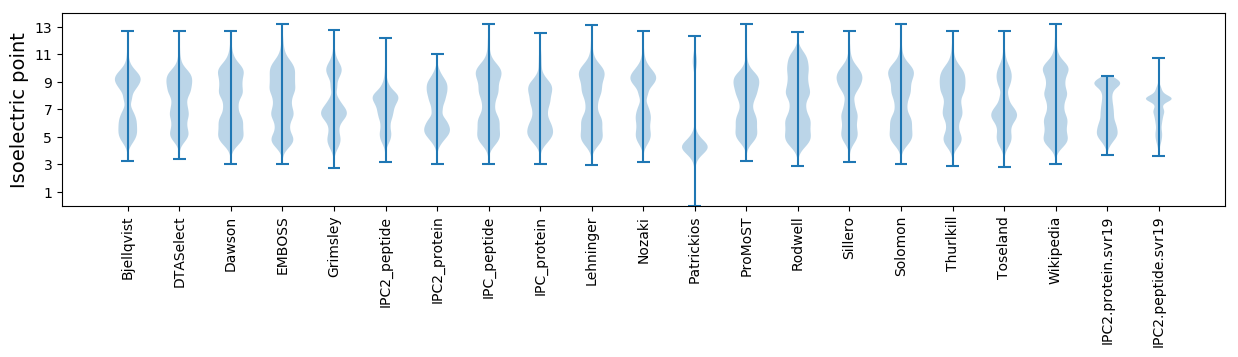
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0L7LQ86|A0A0L7LQ86_9NEOP Zinc/iron transporter OS=Operophtera brumata OX=104452 GN=OBRU01_04041 PE=4 SV=1
MM1 pKa = 7.21VPHH4 pKa = 7.16LPKK7 pKa = 10.43QGPLRR12 pKa = 11.84LNMVPHH18 pKa = 6.81LPKK21 pKa = 10.7LGPLHH26 pKa = 6.9LNMVPHH32 pKa = 6.84LPKK35 pKa = 10.7LGPLHH40 pKa = 6.9LNMVPHH46 pKa = 6.8LPKK49 pKa = 10.81LGTLRR54 pKa = 11.84LNMVPHH60 pKa = 7.02LPKK63 pKa = 10.42QGPLRR68 pKa = 11.84LNMVPHH74 pKa = 6.81LPKK77 pKa = 10.71LGPLRR82 pKa = 11.84LNMVPHH88 pKa = 7.02LPKK91 pKa = 10.42QGPLRR96 pKa = 11.84LNMVFHH102 pKa = 6.88LPKK105 pKa = 10.56LGPFRR110 pKa = 11.84LNMVPHH116 pKa = 6.87LPKK119 pKa = 10.66
MM1 pKa = 7.21VPHH4 pKa = 7.16LPKK7 pKa = 10.43QGPLRR12 pKa = 11.84LNMVPHH18 pKa = 6.81LPKK21 pKa = 10.7LGPLHH26 pKa = 6.9LNMVPHH32 pKa = 6.84LPKK35 pKa = 10.7LGPLHH40 pKa = 6.9LNMVPHH46 pKa = 6.8LPKK49 pKa = 10.81LGTLRR54 pKa = 11.84LNMVPHH60 pKa = 7.02LPKK63 pKa = 10.42QGPLRR68 pKa = 11.84LNMVPHH74 pKa = 6.81LPKK77 pKa = 10.71LGPLRR82 pKa = 11.84LNMVPHH88 pKa = 7.02LPKK91 pKa = 10.42QGPLRR96 pKa = 11.84LNMVFHH102 pKa = 6.88LPKK105 pKa = 10.56LGPFRR110 pKa = 11.84LNMVPHH116 pKa = 6.87LPKK119 pKa = 10.66
Molecular weight: 13.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6022392 |
12 |
16700 |
358.2 |
40.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.03 ± 0.027 | 2.116 ± 0.034 |
5.506 ± 0.016 | 6.469 ± 0.026 |
3.574 ± 0.015 | 5.525 ± 0.024 |
2.624 ± 0.013 | 5.349 ± 0.017 |
6.313 ± 0.027 | 9.111 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.305 ± 0.011 | 4.72 ± 0.019 |
5.317 ± 0.031 | 3.872 ± 0.015 |
5.799 ± 0.025 | 7.727 ± 0.025 |
5.893 ± 0.023 | 6.38 ± 0.018 |
1.122 ± 0.008 | 3.243 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
