
Candidatus Aquiluna sp. IMCC13023
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Luna cluster; Luna-1 subcluster; Aquiluna; unclassified Aquiluna
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
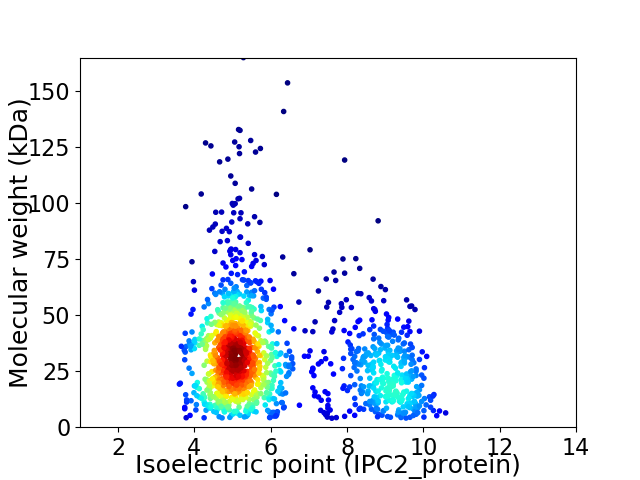
Virtual 2D-PAGE plot for 1364 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I0R1S5|I0R1S5_9MICO Peptidyl-prolyl cis-trans isomerase OS=Candidatus Aquiluna sp. IMCC13023 OX=1081644 GN=IMCC13023_02900 PE=3 SV=1
MM1 pKa = 7.53IKK3 pKa = 10.45SKK5 pKa = 9.4TFSMLAIGATAALVLAGCAAAEE27 pKa = 4.41TEE29 pKa = 4.55TEE31 pKa = 4.3TEE33 pKa = 4.23TTAGTEE39 pKa = 3.81AATSIIGAYY48 pKa = 9.3DD49 pKa = 3.64LSGIDD54 pKa = 3.15ITVGSKK60 pKa = 10.89DD61 pKa = 3.47FDD63 pKa = 3.58EE64 pKa = 4.2QLILGEE70 pKa = 4.03MLVAAFEE77 pKa = 4.32EE78 pKa = 4.66AGATVDD84 pKa = 3.75NKK86 pKa = 10.51VNLGGTSVARR96 pKa = 11.84AALEE100 pKa = 4.23SGDD103 pKa = 2.93IDD105 pKa = 4.8IYY107 pKa = 10.64MEE109 pKa = 4.17YY110 pKa = 10.64NGTGWAVHH118 pKa = 6.77LGQEE122 pKa = 4.66DD123 pKa = 4.3PSFDD127 pKa = 4.18PDD129 pKa = 3.45VLTPEE134 pKa = 4.24VAALDD139 pKa = 3.81LSEE142 pKa = 5.51NGIVWVGRR150 pKa = 11.84SPFNNTYY157 pKa = 10.74GFASSPAVTEE167 pKa = 4.23ANGGAFTLTSMMEE180 pKa = 4.0YY181 pKa = 9.33TQANPDD187 pKa = 4.34AIVCMEE193 pKa = 4.35SEE195 pKa = 4.73FPSRR199 pKa = 11.84PDD201 pKa = 3.56GLILTEE207 pKa = 3.97NHH209 pKa = 6.49SGITLPDD216 pKa = 3.36SQQRR220 pKa = 11.84ILDD223 pKa = 3.3TGIIYY228 pKa = 9.73TEE230 pKa = 4.36TSANNCDD237 pKa = 3.71FGEE240 pKa = 4.72VYY242 pKa = 9.73ATDD245 pKa = 3.43GRR247 pKa = 11.84IPALGLTLVTDD258 pKa = 4.18PGVNILYY265 pKa = 9.94NVSGTIGQDD274 pKa = 3.21KK275 pKa = 10.8YY276 pKa = 11.78NEE278 pKa = 4.34APDD281 pKa = 3.99AFDD284 pKa = 5.78AIINAILAPLTNVRR298 pKa = 11.84MAEE301 pKa = 3.95LNGYY305 pKa = 9.12VSAEE309 pKa = 4.12GQDD312 pKa = 3.45SGVVARR318 pKa = 11.84EE319 pKa = 3.87YY320 pKa = 10.85LVSEE324 pKa = 4.29GLIDD328 pKa = 3.64
MM1 pKa = 7.53IKK3 pKa = 10.45SKK5 pKa = 9.4TFSMLAIGATAALVLAGCAAAEE27 pKa = 4.41TEE29 pKa = 4.55TEE31 pKa = 4.3TEE33 pKa = 4.23TTAGTEE39 pKa = 3.81AATSIIGAYY48 pKa = 9.3DD49 pKa = 3.64LSGIDD54 pKa = 3.15ITVGSKK60 pKa = 10.89DD61 pKa = 3.47FDD63 pKa = 3.58EE64 pKa = 4.2QLILGEE70 pKa = 4.03MLVAAFEE77 pKa = 4.32EE78 pKa = 4.66AGATVDD84 pKa = 3.75NKK86 pKa = 10.51VNLGGTSVARR96 pKa = 11.84AALEE100 pKa = 4.23SGDD103 pKa = 2.93IDD105 pKa = 4.8IYY107 pKa = 10.64MEE109 pKa = 4.17YY110 pKa = 10.64NGTGWAVHH118 pKa = 6.77LGQEE122 pKa = 4.66DD123 pKa = 4.3PSFDD127 pKa = 4.18PDD129 pKa = 3.45VLTPEE134 pKa = 4.24VAALDD139 pKa = 3.81LSEE142 pKa = 5.51NGIVWVGRR150 pKa = 11.84SPFNNTYY157 pKa = 10.74GFASSPAVTEE167 pKa = 4.23ANGGAFTLTSMMEE180 pKa = 4.0YY181 pKa = 9.33TQANPDD187 pKa = 4.34AIVCMEE193 pKa = 4.35SEE195 pKa = 4.73FPSRR199 pKa = 11.84PDD201 pKa = 3.56GLILTEE207 pKa = 3.97NHH209 pKa = 6.49SGITLPDD216 pKa = 3.36SQQRR220 pKa = 11.84ILDD223 pKa = 3.3TGIIYY228 pKa = 9.73TEE230 pKa = 4.36TSANNCDD237 pKa = 3.71FGEE240 pKa = 4.72VYY242 pKa = 9.73ATDD245 pKa = 3.43GRR247 pKa = 11.84IPALGLTLVTDD258 pKa = 4.18PGVNILYY265 pKa = 9.94NVSGTIGQDD274 pKa = 3.21KK275 pKa = 10.8YY276 pKa = 11.78NEE278 pKa = 4.34APDD281 pKa = 3.99AFDD284 pKa = 5.78AIINAILAPLTNVRR298 pKa = 11.84MAEE301 pKa = 3.95LNGYY305 pKa = 9.12VSAEE309 pKa = 4.12GQDD312 pKa = 3.45SGVVARR318 pKa = 11.84EE319 pKa = 3.87YY320 pKa = 10.85LVSEE324 pKa = 4.29GLIDD328 pKa = 3.64
Molecular weight: 34.41 kDa
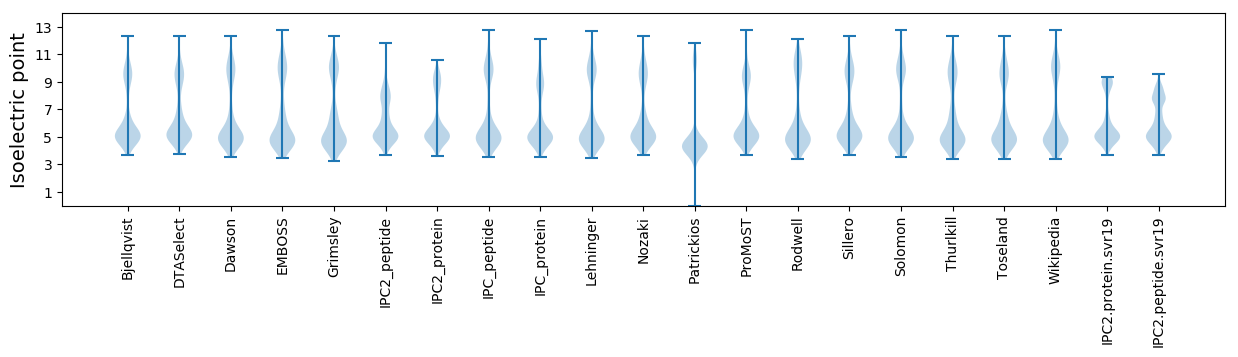
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I0R302|I0R302_9MICO rRNA (Guanine-N2-)-methyltransferase OS=Candidatus Aquiluna sp. IMCC13023 OX=1081644 GN=IMCC13023_07170 PE=4 SV=1
MM1 pKa = 7.89PPRR4 pKa = 11.84LGTSLCSNSSRR15 pKa = 11.84FVLFDD20 pKa = 3.39LRR22 pKa = 11.84DD23 pKa = 4.18FPRR26 pKa = 11.84EE27 pKa = 3.88PPLPNAPAVPAPRR40 pKa = 11.84PPRR43 pKa = 11.84PAGPGLPAGGPNPGFCPGAPNAGAPPGRR71 pKa = 11.84GAPPPGRR78 pKa = 11.84GAPPAGLGAPGRR90 pKa = 11.84GAPGRR95 pKa = 11.84GADD98 pKa = 3.93PAGLGAPGRR107 pKa = 11.84DD108 pKa = 2.78RR109 pKa = 11.84PMPWFAANGLFPGRR123 pKa = 11.84TIGRR127 pKa = 11.84GIPMPWFEE135 pKa = 5.1EE136 pKa = 3.89NGLLPGRR143 pKa = 11.84GVPSIVAAADD153 pKa = 4.17FSSTTGSPASGAEE166 pKa = 3.99AGSGSTATGAAGSVMSEE183 pKa = 4.09GFGLSATAGASGAGLAFGFAGSFLAGASFFGKK215 pKa = 10.54APFSLAATGGVIVEE229 pKa = 4.71EE230 pKa = 4.62PPLTYY235 pKa = 10.51SPSSSNLAKK244 pKa = 10.41AVLVSIPSSLAMSYY258 pKa = 9.63TRR260 pKa = 11.84GSATFFLLFGADD272 pKa = 3.68PKK274 pKa = 10.78QGRR277 pKa = 11.84PP278 pKa = 3.3
MM1 pKa = 7.89PPRR4 pKa = 11.84LGTSLCSNSSRR15 pKa = 11.84FVLFDD20 pKa = 3.39LRR22 pKa = 11.84DD23 pKa = 4.18FPRR26 pKa = 11.84EE27 pKa = 3.88PPLPNAPAVPAPRR40 pKa = 11.84PPRR43 pKa = 11.84PAGPGLPAGGPNPGFCPGAPNAGAPPGRR71 pKa = 11.84GAPPPGRR78 pKa = 11.84GAPPAGLGAPGRR90 pKa = 11.84GAPGRR95 pKa = 11.84GADD98 pKa = 3.93PAGLGAPGRR107 pKa = 11.84DD108 pKa = 2.78RR109 pKa = 11.84PMPWFAANGLFPGRR123 pKa = 11.84TIGRR127 pKa = 11.84GIPMPWFEE135 pKa = 5.1EE136 pKa = 3.89NGLLPGRR143 pKa = 11.84GVPSIVAAADD153 pKa = 4.17FSSTTGSPASGAEE166 pKa = 3.99AGSGSTATGAAGSVMSEE183 pKa = 4.09GFGLSATAGASGAGLAFGFAGSFLAGASFFGKK215 pKa = 10.54APFSLAATGGVIVEE229 pKa = 4.71EE230 pKa = 4.62PPLTYY235 pKa = 10.51SPSSSNLAKK244 pKa = 10.41AVLVSIPSSLAMSYY258 pKa = 9.63TRR260 pKa = 11.84GSATFFLLFGADD272 pKa = 3.68PKK274 pKa = 10.78QGRR277 pKa = 11.84PP278 pKa = 3.3
Molecular weight: 27.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
417606 |
37 |
1517 |
306.2 |
33.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.677 ± 0.078 | 0.537 ± 0.016 |
5.414 ± 0.054 | 6.347 ± 0.074 |
3.806 ± 0.046 | 8.128 ± 0.048 |
1.679 ± 0.03 | 6.065 ± 0.052 |
4.442 ± 0.053 | 10.58 ± 0.088 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.215 ± 0.029 | 3.166 ± 0.033 |
4.334 ± 0.039 | 3.54 ± 0.042 |
5.16 ± 0.046 | 6.997 ± 0.052 |
5.499 ± 0.053 | 8.027 ± 0.065 |
1.197 ± 0.027 | 2.19 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
