
Gramella salexigens
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Gramella
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
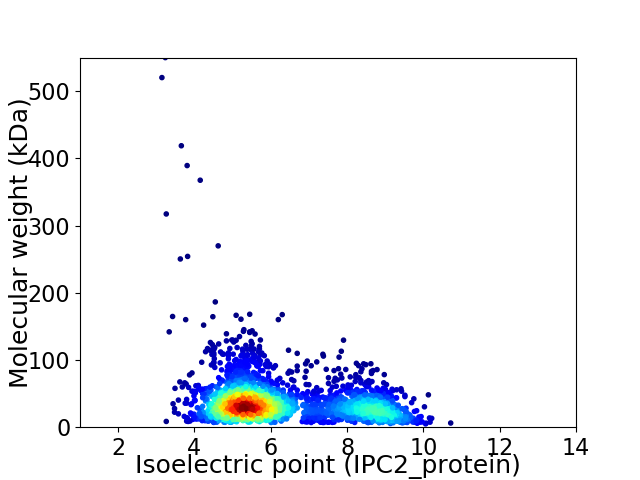
Virtual 2D-PAGE plot for 2591 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3J5G0|A0A1L3J5G0_9FLAO Mechanosensitive ion channel protein OS=Gramella salexigens OX=1913577 GN=LPB144_08150 PE=3 SV=1
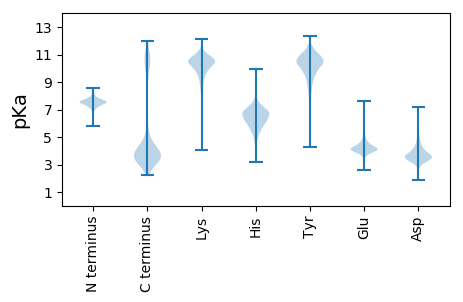
MM1 pKa = 8.1DD2 pKa = 4.13PTRR5 pKa = 11.84DD6 pKa = 3.22VDD8 pKa = 4.09RR9 pKa = 11.84DD10 pKa = 3.5GSPWFIDD17 pKa = 3.93CDD19 pKa = 4.8DD20 pKa = 4.47NNADD24 pKa = 5.34LNDD27 pKa = 3.88KK28 pKa = 11.42DD29 pKa = 4.59EE30 pKa = 6.1DD31 pKa = 3.67NDD33 pKa = 4.48NYY35 pKa = 10.12STCQGDD41 pKa = 5.13PDD43 pKa = 4.18DD44 pKa = 4.72QDD46 pKa = 4.07PNVYY50 pKa = 9.75PGSALICINQTVTAAQTSFICEE72 pKa = 4.11GEE74 pKa = 4.21TTINLGGSQTNVSYY88 pKa = 10.59YY89 pKa = 10.76LRR91 pKa = 11.84NNADD95 pKa = 3.03NSIIDD100 pKa = 3.81GPIAGTGSPISFNTGTISSNTTFNVYY126 pKa = 10.3SEE128 pKa = 4.74AGAKK132 pKa = 9.73GALDD136 pKa = 3.57FDD138 pKa = 5.5GIDD141 pKa = 3.87DD142 pKa = 4.25QVILDD147 pKa = 4.1GFDD150 pKa = 3.53LAGEE154 pKa = 4.15ANVTIEE160 pKa = 3.87AWVNPTDD167 pKa = 3.24VTGAHH172 pKa = 6.86KK173 pKa = 10.9GIISKK178 pKa = 9.45TNGDD182 pKa = 3.41GQFIFRR188 pKa = 11.84LEE190 pKa = 4.17NGHH193 pKa = 6.51PRR195 pKa = 11.84SVFYY199 pKa = 10.62TSTGDD204 pKa = 3.53LGSLSTQALTPGQWYY219 pKa = 9.43HH220 pKa = 6.0VVTTFDD226 pKa = 3.55GDD228 pKa = 3.3EE229 pKa = 3.86SRR231 pKa = 11.84IYY233 pKa = 11.03VNGVDD238 pKa = 3.89VTGAQYY244 pKa = 11.33NYY246 pKa = 10.2AASGGLKK253 pKa = 9.67TYY255 pKa = 7.47TASPAIIGRR264 pKa = 11.84ANPQEE269 pKa = 4.13FFDD272 pKa = 4.07GKK274 pKa = 10.04IGEE277 pKa = 4.11VRR279 pKa = 11.84VWNEE283 pKa = 3.47LKK285 pKa = 10.08TPAEE289 pKa = 3.83ITANMSSSYY298 pKa = 10.5TGTEE302 pKa = 3.69TGLVALYY309 pKa = 10.65KK310 pKa = 9.65MKK312 pKa = 10.08EE313 pKa = 4.13GKK315 pKa = 10.46GITVADD321 pKa = 3.73STGSFDD327 pKa = 3.69GTMEE331 pKa = 4.1NMDD334 pKa = 4.95PATDD338 pKa = 3.38WLTYY342 pKa = 10.92DD343 pKa = 4.18PDD345 pKa = 3.95CTLEE349 pKa = 4.15MTDD352 pKa = 4.15KK353 pKa = 10.7PSVTIAPIEE362 pKa = 4.27DD363 pKa = 3.44QVVTVAEE370 pKa = 4.4STLCPSNTGTTVNLEE385 pKa = 4.02TSQPDD390 pKa = 2.87ITYY393 pKa = 10.55YY394 pKa = 10.75LRR396 pKa = 11.84NDD398 pKa = 3.85ADD400 pKa = 4.21DD401 pKa = 5.1SIIDD405 pKa = 3.86GPVEE409 pKa = 4.03GGGAISFDD417 pKa = 3.54TGSITTTTNYY427 pKa = 10.0NVHH430 pKa = 6.78AVPTNGDD437 pKa = 3.9TSCSTQMSQTVAVTIEE453 pKa = 4.82DD454 pKa = 3.52NTAPTVIARR463 pKa = 11.84DD464 pKa = 3.28ITVQLDD470 pKa = 3.52EE471 pKa = 5.15NGQATITADD480 pKa = 3.31QVNNGSTDD488 pKa = 2.81NCTAIGNLTLSLDD501 pKa = 3.56KK502 pKa = 10.97STFSCADD509 pKa = 2.84ISSGGEE515 pKa = 3.87CGEE518 pKa = 4.01VSLLFNSSGIATGDD532 pKa = 3.09NNANVEE538 pKa = 4.26EE539 pKa = 4.84LGMNQLTMEE548 pKa = 4.48AWVKK552 pKa = 10.35PSSNVVNSVIRR563 pKa = 11.84KK564 pKa = 9.52AGDD567 pKa = 3.25YY568 pKa = 10.97DD569 pKa = 4.4LYY571 pKa = 10.69IYY573 pKa = 10.38DD574 pKa = 4.18GQLKK578 pKa = 10.76AEE580 pKa = 4.19VWYY583 pKa = 10.31KK584 pKa = 10.88AGSDD588 pKa = 2.79ASMYY592 pKa = 10.63QFTGPAITYY601 pKa = 7.31NAWSHH606 pKa = 5.05VAFVYY611 pKa = 10.85DD612 pKa = 3.49NTNPGVGYY620 pKa = 9.23FVVNGNTYY628 pKa = 9.68PSNGNAHH635 pKa = 6.31QIGSNGNLGIGSSTVYY651 pKa = 10.16GQSFNGLIDD660 pKa = 3.61EE661 pKa = 4.2VRR663 pKa = 11.84IWSVARR669 pKa = 11.84TAQNIQDD676 pKa = 4.18NLSGCITGSEE686 pKa = 4.34LGLEE690 pKa = 4.46AYY692 pKa = 10.53YY693 pKa = 10.41KK694 pKa = 9.38VQEE697 pKa = 4.38GSGSVLKK704 pKa = 10.87DD705 pKa = 3.68FSDD708 pKa = 3.61NGNDD712 pKa = 3.63LTIQGATWQQEE723 pKa = 4.12GGTTAGVPVTLTVGDD738 pKa = 4.06ANGNQSTATANVAVVDD754 pKa = 5.14IIAPIIRR761 pKa = 11.84TYY763 pKa = 11.44EE764 pKa = 4.7DD765 pKa = 3.13ILVEE769 pKa = 4.28LDD771 pKa = 3.57EE772 pKa = 5.15NGQASIRR779 pKa = 11.84GDD781 pKa = 3.5YY782 pKa = 10.89SVGPVAGKK790 pKa = 10.41LEE792 pKa = 4.29PQNSLSTTTPVDD804 pKa = 3.48CDD806 pKa = 3.63CPEE809 pKa = 4.44GYY811 pKa = 10.26VAVGYY816 pKa = 9.9EE817 pKa = 4.46GASGWILDD825 pKa = 4.74DD826 pKa = 3.97FRR828 pKa = 11.84LVCKK832 pKa = 10.17EE833 pKa = 3.67VLADD837 pKa = 3.84GGLGTEE843 pKa = 4.38TVEE846 pKa = 4.41TCFSGSRR853 pKa = 11.84TEE855 pKa = 4.07VTRR858 pKa = 11.84SDD860 pKa = 3.45MLTGNEE866 pKa = 3.96VLVGFEE872 pKa = 4.12VQDD875 pKa = 4.35GDD877 pKa = 4.12FQHH880 pKa = 6.18QLSSRR885 pKa = 11.84THH887 pKa = 6.02VGVKK891 pKa = 10.56GFGKK895 pKa = 10.57SLDD898 pKa = 3.48QVVAGNQNNIDD909 pKa = 3.65NTALAGIIGSGTGTTSLATTTNFAPAGHH937 pKa = 7.39AIVGMSVNQTSGYY950 pKa = 8.97SSSVSFKK957 pKa = 10.15YY958 pKa = 10.79APISSLLSIDD968 pKa = 4.08NGSTDD973 pKa = 3.0NCGIASITADD983 pKa = 3.43KK984 pKa = 11.49YY985 pKa = 11.56SFTCEE990 pKa = 3.9NIGEE994 pKa = 4.37NIVTITAMDD1003 pKa = 3.63VNGNEE1008 pKa = 4.01ASATAVVTVEE1018 pKa = 5.57DD1019 pKa = 3.68NTAPQAIVRR1028 pKa = 11.84NITVEE1033 pKa = 3.83LDD1035 pKa = 3.43ANGMASITPEE1045 pKa = 3.89MVDD1048 pKa = 3.62NGSSDD1053 pKa = 3.44NCATTEE1059 pKa = 3.86DD1060 pKa = 4.02LQLILDD1066 pKa = 4.0KK1067 pKa = 11.25SIFGCEE1073 pKa = 3.82DD1074 pKa = 3.32VVDD1077 pKa = 4.55KK1078 pKa = 11.33GVTPNLSALNFDD1090 pKa = 3.48GDD1092 pKa = 4.68DD1093 pKa = 3.69YY1094 pKa = 12.07VSIPDD1099 pKa = 3.69SPEE1102 pKa = 3.73LQLQGDD1108 pKa = 4.06MTIEE1112 pKa = 3.46AWFKK1116 pKa = 10.53VDD1118 pKa = 4.42EE1119 pKa = 4.56FTGDD1123 pKa = 2.95WVRR1126 pKa = 11.84VVGKK1130 pKa = 10.1GAGPEE1135 pKa = 4.41GPVGPRR1141 pKa = 11.84NYY1143 pKa = 10.58GLWYY1147 pKa = 9.82FPNGTWLFQQYY1158 pKa = 9.63GQGVQVAFNRR1168 pKa = 11.84PIDD1171 pKa = 3.57IGEE1174 pKa = 4.35WYY1176 pKa = 10.62HH1177 pKa = 5.28MAAVKK1182 pKa = 9.43TGNVAKK1188 pKa = 10.5LYY1190 pKa = 10.55INGEE1194 pKa = 4.41VVASNTGGTNPITSNDD1210 pKa = 3.82PLTIGYY1216 pKa = 10.28AGFHH1220 pKa = 6.66DD1221 pKa = 4.0HH1222 pKa = 7.5HH1223 pKa = 8.08IGQIDD1228 pKa = 4.13EE1229 pKa = 4.05VRR1231 pKa = 11.84LWNIARR1237 pKa = 11.84TDD1239 pKa = 3.67EE1240 pKa = 4.91EE1241 pKa = 4.33ILNDD1245 pKa = 3.43FTKK1248 pKa = 10.44TINPEE1253 pKa = 3.84TSGLLAYY1260 pKa = 11.03YY1261 pKa = 10.82NMEE1264 pKa = 4.23EE1265 pKa = 4.16ASGDD1269 pKa = 3.5ILNDD1273 pKa = 3.12QTVNGFNGDD1282 pKa = 3.46LQGFSQNEE1290 pKa = 3.72AWTGSSRR1297 pKa = 11.84RR1298 pKa = 11.84LGPVSTMLTVTDD1310 pKa = 3.99ASGNSSSSTAVVTVVDD1326 pKa = 5.05NIAATVVTQNITVALDD1342 pKa = 3.54EE1343 pKa = 5.2NGTATITPEE1352 pKa = 4.22MIDD1355 pKa = 3.82GGSSDD1360 pKa = 3.51NCEE1363 pKa = 3.93IASLEE1368 pKa = 4.02LDD1370 pKa = 3.54VTEE1373 pKa = 5.0FTCEE1377 pKa = 3.74NVGEE1381 pKa = 4.25NTVTLTAVDD1390 pKa = 3.93VNDD1393 pKa = 4.09NEE1395 pKa = 4.42SSATAVVTVEE1405 pKa = 4.14DD1406 pKa = 4.56NIPAEE1411 pKa = 4.2VLTQNITVQLDD1422 pKa = 3.6DD1423 pKa = 4.38MGMASITPEE1432 pKa = 4.12MIDD1435 pKa = 3.16NGSNDD1440 pKa = 3.21VCGIASMSLDD1450 pKa = 3.0ITDD1453 pKa = 4.58FTCEE1457 pKa = 3.85NVGEE1461 pKa = 4.14NMVTLTAVDD1470 pKa = 3.86VNDD1473 pKa = 4.09NEE1475 pKa = 4.38ASATATVIVEE1485 pKa = 4.21DD1486 pKa = 4.66NIAATVVTQNITVALDD1502 pKa = 3.54EE1503 pKa = 5.2NGTATITPEE1512 pKa = 4.17MIDD1515 pKa = 3.21NGSNDD1520 pKa = 3.16VCGIASLSLDD1530 pKa = 3.08ITDD1533 pKa = 4.57FTCEE1537 pKa = 3.84NVGEE1541 pKa = 4.25NTVTLTAVDD1550 pKa = 3.95VNDD1553 pKa = 4.09NEE1555 pKa = 4.38ASATATVIVEE1565 pKa = 4.15DD1566 pKa = 4.53KK1567 pKa = 10.68IAAEE1571 pKa = 4.1VLTQNITVQLDD1582 pKa = 3.59EE1583 pKa = 4.93MGMASITPEE1592 pKa = 4.21MIDD1595 pKa = 3.25NDD1597 pKa = 4.33SNDD1600 pKa = 3.24VCGIASLSLDD1610 pKa = 3.08ITDD1613 pKa = 4.57FTCEE1617 pKa = 3.84NVGEE1621 pKa = 4.25NTVTLTAVDD1630 pKa = 3.95VNDD1633 pKa = 4.09NEE1635 pKa = 4.38ASATATVIVEE1645 pKa = 4.15DD1646 pKa = 4.53KK1647 pKa = 10.68IAAEE1651 pKa = 4.1VLTQNITVQLDD1662 pKa = 3.76EE1663 pKa = 6.11LGMASITPEE1672 pKa = 4.12MIDD1675 pKa = 3.37NGSNDD1680 pKa = 3.24ACGIASMSLDD1690 pKa = 3.0ITDD1693 pKa = 4.58FTCEE1697 pKa = 3.84NVGEE1701 pKa = 4.25NTVTLTAVDD1710 pKa = 3.95VNDD1713 pKa = 4.09NEE1715 pKa = 4.38ASATATVIVEE1725 pKa = 4.15DD1726 pKa = 4.53KK1727 pKa = 10.68IAAEE1731 pKa = 4.1VLTQNITVQLDD1742 pKa = 3.6DD1743 pKa = 4.38MGMASITPEE1752 pKa = 4.02MINNGSNDD1760 pKa = 3.2VCGIASMSLDD1770 pKa = 3.0ITDD1773 pKa = 4.58FTCEE1777 pKa = 3.84NVGEE1781 pKa = 4.27NTVTLTVVDD1790 pKa = 4.24VNDD1793 pKa = 3.89NEE1795 pKa = 4.37ASATATVIVEE1805 pKa = 4.15DD1806 pKa = 4.53KK1807 pKa = 10.68IAAEE1811 pKa = 4.1VLTQNITVQLDD1822 pKa = 3.59EE1823 pKa = 4.93MGMASITPEE1832 pKa = 4.12MIDD1835 pKa = 3.37NGSNDD1840 pKa = 3.24ACGIASMSLDD1850 pKa = 3.0ITDD1853 pKa = 4.58FTCEE1857 pKa = 3.85NVGEE1861 pKa = 4.36NIVTLTAMDD1870 pKa = 3.84VNDD1873 pKa = 4.17NEE1875 pKa = 4.47ASATVIVTVEE1885 pKa = 3.97DD1886 pKa = 4.05KK1887 pKa = 10.84IAAEE1891 pKa = 4.1VLTQNITVQLDD1902 pKa = 3.59EE1903 pKa = 4.93MGMASITPEE1912 pKa = 4.12MIDD1915 pKa = 3.37NGSNDD1920 pKa = 2.95ACGIVSMSLDD1930 pKa = 3.15ITDD1933 pKa = 4.58FTCEE1937 pKa = 3.85NVGEE1941 pKa = 4.36NIVTLTAMDD1950 pKa = 3.84VNDD1953 pKa = 4.13NEE1955 pKa = 4.37ASATAIVTVEE1965 pKa = 4.06DD1966 pKa = 4.18KK1967 pKa = 10.84IAAEE1971 pKa = 4.47VITQNITVQLDD1982 pKa = 3.59EE1983 pKa = 4.93MGMASITPEE1992 pKa = 4.12MIDD1995 pKa = 3.37NGSNDD2000 pKa = 3.24ACGIASMSLDD2010 pKa = 3.69KK2011 pKa = 11.4SDD2013 pKa = 4.16FTCDD2017 pKa = 3.34HH2018 pKa = 6.62IGEE2021 pKa = 4.34NTVTLTAIDD2030 pKa = 3.21IHH2032 pKa = 8.19GNEE2035 pKa = 4.25SSATAIVSVEE2045 pKa = 4.09DD2046 pKa = 4.08VIAPTPVMDD2055 pKa = 3.88QLEE2058 pKa = 4.84PIYY2061 pKa = 10.64AEE2063 pKa = 4.52CIVEE2067 pKa = 4.21RR2068 pKa = 11.84TDD2070 pKa = 3.55VPTPLATDD2078 pKa = 3.2NCDD2081 pKa = 3.15IEE2083 pKa = 5.31ISATTDD2089 pKa = 3.1LVFPVTRR2096 pKa = 11.84QGSTMIRR2103 pKa = 11.84WKK2105 pKa = 11.03FEE2107 pKa = 4.03DD2108 pKa = 3.34SNANVTYY2115 pKa = 9.6QQQKK2119 pKa = 10.46IIIDD2123 pKa = 4.96DD2124 pKa = 3.44ITAPVPDD2131 pKa = 3.65IAEE2134 pKa = 4.38LEE2136 pKa = 4.42TVEE2139 pKa = 4.42VEE2141 pKa = 4.44CGVTAISAPTATDD2154 pKa = 2.93NCAGSVTGTTTDD2166 pKa = 3.45PLSYY2170 pKa = 10.45EE2171 pKa = 3.92EE2172 pKa = 4.07QGDD2175 pKa = 4.2YY2176 pKa = 11.0EE2177 pKa = 4.35ILWTFDD2183 pKa = 3.57DD2184 pKa = 4.83GNGNSTTQIQQVIVKK2199 pKa = 9.71DD2200 pKa = 3.47QTPPVALAKK2209 pKa = 10.52DD2210 pKa = 3.36ITVVLDD2216 pKa = 3.64PNDD2219 pKa = 3.39NVRR2222 pKa = 11.84INPEE2226 pKa = 4.49DD2227 pKa = 4.1IDD2229 pKa = 4.28DD2230 pKa = 4.25GSYY2233 pKa = 11.07DD2234 pKa = 3.65EE2235 pKa = 5.3CSEE2238 pKa = 4.08ISLSLDD2244 pKa = 2.86RR2245 pKa = 11.84DD2246 pKa = 4.01YY2247 pKa = 10.87FTSNGSYY2254 pKa = 10.23EE2255 pKa = 4.37VVLTVTDD2262 pKa = 3.83ALGNSSEE2269 pKa = 3.84ASARR2273 pKa = 11.84VIVVTNHH2280 pKa = 6.75PEE2282 pKa = 3.76TSNVHH2287 pKa = 4.77VVPTILKK2294 pKa = 7.92QSSMAKK2300 pKa = 9.59VIVPFRR2306 pKa = 11.84SRR2308 pKa = 11.84IIEE2311 pKa = 4.14VQVLEE2316 pKa = 4.45TEE2318 pKa = 4.22TNKK2321 pKa = 10.62YY2322 pKa = 10.31KK2323 pKa = 10.62IIRR2326 pKa = 11.84GNKK2329 pKa = 9.57LNEE2332 pKa = 3.89QQIDD2336 pKa = 3.33IAPFKK2341 pKa = 10.18GTLLVRR2347 pKa = 11.84IIDD2350 pKa = 3.47QDD2352 pKa = 3.65GRR2354 pKa = 11.84VHH2356 pKa = 6.86LKK2358 pKa = 10.68KK2359 pKa = 10.9LIALL2363 pKa = 4.36
MM1 pKa = 8.1DD2 pKa = 4.13PTRR5 pKa = 11.84DD6 pKa = 3.22VDD8 pKa = 4.09RR9 pKa = 11.84DD10 pKa = 3.5GSPWFIDD17 pKa = 3.93CDD19 pKa = 4.8DD20 pKa = 4.47NNADD24 pKa = 5.34LNDD27 pKa = 3.88KK28 pKa = 11.42DD29 pKa = 4.59EE30 pKa = 6.1DD31 pKa = 3.67NDD33 pKa = 4.48NYY35 pKa = 10.12STCQGDD41 pKa = 5.13PDD43 pKa = 4.18DD44 pKa = 4.72QDD46 pKa = 4.07PNVYY50 pKa = 9.75PGSALICINQTVTAAQTSFICEE72 pKa = 4.11GEE74 pKa = 4.21TTINLGGSQTNVSYY88 pKa = 10.59YY89 pKa = 10.76LRR91 pKa = 11.84NNADD95 pKa = 3.03NSIIDD100 pKa = 3.81GPIAGTGSPISFNTGTISSNTTFNVYY126 pKa = 10.3SEE128 pKa = 4.74AGAKK132 pKa = 9.73GALDD136 pKa = 3.57FDD138 pKa = 5.5GIDD141 pKa = 3.87DD142 pKa = 4.25QVILDD147 pKa = 4.1GFDD150 pKa = 3.53LAGEE154 pKa = 4.15ANVTIEE160 pKa = 3.87AWVNPTDD167 pKa = 3.24VTGAHH172 pKa = 6.86KK173 pKa = 10.9GIISKK178 pKa = 9.45TNGDD182 pKa = 3.41GQFIFRR188 pKa = 11.84LEE190 pKa = 4.17NGHH193 pKa = 6.51PRR195 pKa = 11.84SVFYY199 pKa = 10.62TSTGDD204 pKa = 3.53LGSLSTQALTPGQWYY219 pKa = 9.43HH220 pKa = 6.0VVTTFDD226 pKa = 3.55GDD228 pKa = 3.3EE229 pKa = 3.86SRR231 pKa = 11.84IYY233 pKa = 11.03VNGVDD238 pKa = 3.89VTGAQYY244 pKa = 11.33NYY246 pKa = 10.2AASGGLKK253 pKa = 9.67TYY255 pKa = 7.47TASPAIIGRR264 pKa = 11.84ANPQEE269 pKa = 4.13FFDD272 pKa = 4.07GKK274 pKa = 10.04IGEE277 pKa = 4.11VRR279 pKa = 11.84VWNEE283 pKa = 3.47LKK285 pKa = 10.08TPAEE289 pKa = 3.83ITANMSSSYY298 pKa = 10.5TGTEE302 pKa = 3.69TGLVALYY309 pKa = 10.65KK310 pKa = 9.65MKK312 pKa = 10.08EE313 pKa = 4.13GKK315 pKa = 10.46GITVADD321 pKa = 3.73STGSFDD327 pKa = 3.69GTMEE331 pKa = 4.1NMDD334 pKa = 4.95PATDD338 pKa = 3.38WLTYY342 pKa = 10.92DD343 pKa = 4.18PDD345 pKa = 3.95CTLEE349 pKa = 4.15MTDD352 pKa = 4.15KK353 pKa = 10.7PSVTIAPIEE362 pKa = 4.27DD363 pKa = 3.44QVVTVAEE370 pKa = 4.4STLCPSNTGTTVNLEE385 pKa = 4.02TSQPDD390 pKa = 2.87ITYY393 pKa = 10.55YY394 pKa = 10.75LRR396 pKa = 11.84NDD398 pKa = 3.85ADD400 pKa = 4.21DD401 pKa = 5.1SIIDD405 pKa = 3.86GPVEE409 pKa = 4.03GGGAISFDD417 pKa = 3.54TGSITTTTNYY427 pKa = 10.0NVHH430 pKa = 6.78AVPTNGDD437 pKa = 3.9TSCSTQMSQTVAVTIEE453 pKa = 4.82DD454 pKa = 3.52NTAPTVIARR463 pKa = 11.84DD464 pKa = 3.28ITVQLDD470 pKa = 3.52EE471 pKa = 5.15NGQATITADD480 pKa = 3.31QVNNGSTDD488 pKa = 2.81NCTAIGNLTLSLDD501 pKa = 3.56KK502 pKa = 10.97STFSCADD509 pKa = 2.84ISSGGEE515 pKa = 3.87CGEE518 pKa = 4.01VSLLFNSSGIATGDD532 pKa = 3.09NNANVEE538 pKa = 4.26EE539 pKa = 4.84LGMNQLTMEE548 pKa = 4.48AWVKK552 pKa = 10.35PSSNVVNSVIRR563 pKa = 11.84KK564 pKa = 9.52AGDD567 pKa = 3.25YY568 pKa = 10.97DD569 pKa = 4.4LYY571 pKa = 10.69IYY573 pKa = 10.38DD574 pKa = 4.18GQLKK578 pKa = 10.76AEE580 pKa = 4.19VWYY583 pKa = 10.31KK584 pKa = 10.88AGSDD588 pKa = 2.79ASMYY592 pKa = 10.63QFTGPAITYY601 pKa = 7.31NAWSHH606 pKa = 5.05VAFVYY611 pKa = 10.85DD612 pKa = 3.49NTNPGVGYY620 pKa = 9.23FVVNGNTYY628 pKa = 9.68PSNGNAHH635 pKa = 6.31QIGSNGNLGIGSSTVYY651 pKa = 10.16GQSFNGLIDD660 pKa = 3.61EE661 pKa = 4.2VRR663 pKa = 11.84IWSVARR669 pKa = 11.84TAQNIQDD676 pKa = 4.18NLSGCITGSEE686 pKa = 4.34LGLEE690 pKa = 4.46AYY692 pKa = 10.53YY693 pKa = 10.41KK694 pKa = 9.38VQEE697 pKa = 4.38GSGSVLKK704 pKa = 10.87DD705 pKa = 3.68FSDD708 pKa = 3.61NGNDD712 pKa = 3.63LTIQGATWQQEE723 pKa = 4.12GGTTAGVPVTLTVGDD738 pKa = 4.06ANGNQSTATANVAVVDD754 pKa = 5.14IIAPIIRR761 pKa = 11.84TYY763 pKa = 11.44EE764 pKa = 4.7DD765 pKa = 3.13ILVEE769 pKa = 4.28LDD771 pKa = 3.57EE772 pKa = 5.15NGQASIRR779 pKa = 11.84GDD781 pKa = 3.5YY782 pKa = 10.89SVGPVAGKK790 pKa = 10.41LEE792 pKa = 4.29PQNSLSTTTPVDD804 pKa = 3.48CDD806 pKa = 3.63CPEE809 pKa = 4.44GYY811 pKa = 10.26VAVGYY816 pKa = 9.9EE817 pKa = 4.46GASGWILDD825 pKa = 4.74DD826 pKa = 3.97FRR828 pKa = 11.84LVCKK832 pKa = 10.17EE833 pKa = 3.67VLADD837 pKa = 3.84GGLGTEE843 pKa = 4.38TVEE846 pKa = 4.41TCFSGSRR853 pKa = 11.84TEE855 pKa = 4.07VTRR858 pKa = 11.84SDD860 pKa = 3.45MLTGNEE866 pKa = 3.96VLVGFEE872 pKa = 4.12VQDD875 pKa = 4.35GDD877 pKa = 4.12FQHH880 pKa = 6.18QLSSRR885 pKa = 11.84THH887 pKa = 6.02VGVKK891 pKa = 10.56GFGKK895 pKa = 10.57SLDD898 pKa = 3.48QVVAGNQNNIDD909 pKa = 3.65NTALAGIIGSGTGTTSLATTTNFAPAGHH937 pKa = 7.39AIVGMSVNQTSGYY950 pKa = 8.97SSSVSFKK957 pKa = 10.15YY958 pKa = 10.79APISSLLSIDD968 pKa = 4.08NGSTDD973 pKa = 3.0NCGIASITADD983 pKa = 3.43KK984 pKa = 11.49YY985 pKa = 11.56SFTCEE990 pKa = 3.9NIGEE994 pKa = 4.37NIVTITAMDD1003 pKa = 3.63VNGNEE1008 pKa = 4.01ASATAVVTVEE1018 pKa = 5.57DD1019 pKa = 3.68NTAPQAIVRR1028 pKa = 11.84NITVEE1033 pKa = 3.83LDD1035 pKa = 3.43ANGMASITPEE1045 pKa = 3.89MVDD1048 pKa = 3.62NGSSDD1053 pKa = 3.44NCATTEE1059 pKa = 3.86DD1060 pKa = 4.02LQLILDD1066 pKa = 4.0KK1067 pKa = 11.25SIFGCEE1073 pKa = 3.82DD1074 pKa = 3.32VVDD1077 pKa = 4.55KK1078 pKa = 11.33GVTPNLSALNFDD1090 pKa = 3.48GDD1092 pKa = 4.68DD1093 pKa = 3.69YY1094 pKa = 12.07VSIPDD1099 pKa = 3.69SPEE1102 pKa = 3.73LQLQGDD1108 pKa = 4.06MTIEE1112 pKa = 3.46AWFKK1116 pKa = 10.53VDD1118 pKa = 4.42EE1119 pKa = 4.56FTGDD1123 pKa = 2.95WVRR1126 pKa = 11.84VVGKK1130 pKa = 10.1GAGPEE1135 pKa = 4.41GPVGPRR1141 pKa = 11.84NYY1143 pKa = 10.58GLWYY1147 pKa = 9.82FPNGTWLFQQYY1158 pKa = 9.63GQGVQVAFNRR1168 pKa = 11.84PIDD1171 pKa = 3.57IGEE1174 pKa = 4.35WYY1176 pKa = 10.62HH1177 pKa = 5.28MAAVKK1182 pKa = 9.43TGNVAKK1188 pKa = 10.5LYY1190 pKa = 10.55INGEE1194 pKa = 4.41VVASNTGGTNPITSNDD1210 pKa = 3.82PLTIGYY1216 pKa = 10.28AGFHH1220 pKa = 6.66DD1221 pKa = 4.0HH1222 pKa = 7.5HH1223 pKa = 8.08IGQIDD1228 pKa = 4.13EE1229 pKa = 4.05VRR1231 pKa = 11.84LWNIARR1237 pKa = 11.84TDD1239 pKa = 3.67EE1240 pKa = 4.91EE1241 pKa = 4.33ILNDD1245 pKa = 3.43FTKK1248 pKa = 10.44TINPEE1253 pKa = 3.84TSGLLAYY1260 pKa = 11.03YY1261 pKa = 10.82NMEE1264 pKa = 4.23EE1265 pKa = 4.16ASGDD1269 pKa = 3.5ILNDD1273 pKa = 3.12QTVNGFNGDD1282 pKa = 3.46LQGFSQNEE1290 pKa = 3.72AWTGSSRR1297 pKa = 11.84RR1298 pKa = 11.84LGPVSTMLTVTDD1310 pKa = 3.99ASGNSSSSTAVVTVVDD1326 pKa = 5.05NIAATVVTQNITVALDD1342 pKa = 3.54EE1343 pKa = 5.2NGTATITPEE1352 pKa = 4.22MIDD1355 pKa = 3.82GGSSDD1360 pKa = 3.51NCEE1363 pKa = 3.93IASLEE1368 pKa = 4.02LDD1370 pKa = 3.54VTEE1373 pKa = 5.0FTCEE1377 pKa = 3.74NVGEE1381 pKa = 4.25NTVTLTAVDD1390 pKa = 3.93VNDD1393 pKa = 4.09NEE1395 pKa = 4.42SSATAVVTVEE1405 pKa = 4.14DD1406 pKa = 4.56NIPAEE1411 pKa = 4.2VLTQNITVQLDD1422 pKa = 3.6DD1423 pKa = 4.38MGMASITPEE1432 pKa = 4.12MIDD1435 pKa = 3.16NGSNDD1440 pKa = 3.21VCGIASMSLDD1450 pKa = 3.0ITDD1453 pKa = 4.58FTCEE1457 pKa = 3.85NVGEE1461 pKa = 4.14NMVTLTAVDD1470 pKa = 3.86VNDD1473 pKa = 4.09NEE1475 pKa = 4.38ASATATVIVEE1485 pKa = 4.21DD1486 pKa = 4.66NIAATVVTQNITVALDD1502 pKa = 3.54EE1503 pKa = 5.2NGTATITPEE1512 pKa = 4.17MIDD1515 pKa = 3.21NGSNDD1520 pKa = 3.16VCGIASLSLDD1530 pKa = 3.08ITDD1533 pKa = 4.57FTCEE1537 pKa = 3.84NVGEE1541 pKa = 4.25NTVTLTAVDD1550 pKa = 3.95VNDD1553 pKa = 4.09NEE1555 pKa = 4.38ASATATVIVEE1565 pKa = 4.15DD1566 pKa = 4.53KK1567 pKa = 10.68IAAEE1571 pKa = 4.1VLTQNITVQLDD1582 pKa = 3.59EE1583 pKa = 4.93MGMASITPEE1592 pKa = 4.21MIDD1595 pKa = 3.25NDD1597 pKa = 4.33SNDD1600 pKa = 3.24VCGIASLSLDD1610 pKa = 3.08ITDD1613 pKa = 4.57FTCEE1617 pKa = 3.84NVGEE1621 pKa = 4.25NTVTLTAVDD1630 pKa = 3.95VNDD1633 pKa = 4.09NEE1635 pKa = 4.38ASATATVIVEE1645 pKa = 4.15DD1646 pKa = 4.53KK1647 pKa = 10.68IAAEE1651 pKa = 4.1VLTQNITVQLDD1662 pKa = 3.76EE1663 pKa = 6.11LGMASITPEE1672 pKa = 4.12MIDD1675 pKa = 3.37NGSNDD1680 pKa = 3.24ACGIASMSLDD1690 pKa = 3.0ITDD1693 pKa = 4.58FTCEE1697 pKa = 3.84NVGEE1701 pKa = 4.25NTVTLTAVDD1710 pKa = 3.95VNDD1713 pKa = 4.09NEE1715 pKa = 4.38ASATATVIVEE1725 pKa = 4.15DD1726 pKa = 4.53KK1727 pKa = 10.68IAAEE1731 pKa = 4.1VLTQNITVQLDD1742 pKa = 3.6DD1743 pKa = 4.38MGMASITPEE1752 pKa = 4.02MINNGSNDD1760 pKa = 3.2VCGIASMSLDD1770 pKa = 3.0ITDD1773 pKa = 4.58FTCEE1777 pKa = 3.84NVGEE1781 pKa = 4.27NTVTLTVVDD1790 pKa = 4.24VNDD1793 pKa = 3.89NEE1795 pKa = 4.37ASATATVIVEE1805 pKa = 4.15DD1806 pKa = 4.53KK1807 pKa = 10.68IAAEE1811 pKa = 4.1VLTQNITVQLDD1822 pKa = 3.59EE1823 pKa = 4.93MGMASITPEE1832 pKa = 4.12MIDD1835 pKa = 3.37NGSNDD1840 pKa = 3.24ACGIASMSLDD1850 pKa = 3.0ITDD1853 pKa = 4.58FTCEE1857 pKa = 3.85NVGEE1861 pKa = 4.36NIVTLTAMDD1870 pKa = 3.84VNDD1873 pKa = 4.17NEE1875 pKa = 4.47ASATVIVTVEE1885 pKa = 3.97DD1886 pKa = 4.05KK1887 pKa = 10.84IAAEE1891 pKa = 4.1VLTQNITVQLDD1902 pKa = 3.59EE1903 pKa = 4.93MGMASITPEE1912 pKa = 4.12MIDD1915 pKa = 3.37NGSNDD1920 pKa = 2.95ACGIVSMSLDD1930 pKa = 3.15ITDD1933 pKa = 4.58FTCEE1937 pKa = 3.85NVGEE1941 pKa = 4.36NIVTLTAMDD1950 pKa = 3.84VNDD1953 pKa = 4.13NEE1955 pKa = 4.37ASATAIVTVEE1965 pKa = 4.06DD1966 pKa = 4.18KK1967 pKa = 10.84IAAEE1971 pKa = 4.47VITQNITVQLDD1982 pKa = 3.59EE1983 pKa = 4.93MGMASITPEE1992 pKa = 4.12MIDD1995 pKa = 3.37NGSNDD2000 pKa = 3.24ACGIASMSLDD2010 pKa = 3.69KK2011 pKa = 11.4SDD2013 pKa = 4.16FTCDD2017 pKa = 3.34HH2018 pKa = 6.62IGEE2021 pKa = 4.34NTVTLTAIDD2030 pKa = 3.21IHH2032 pKa = 8.19GNEE2035 pKa = 4.25SSATAIVSVEE2045 pKa = 4.09DD2046 pKa = 4.08VIAPTPVMDD2055 pKa = 3.88QLEE2058 pKa = 4.84PIYY2061 pKa = 10.64AEE2063 pKa = 4.52CIVEE2067 pKa = 4.21RR2068 pKa = 11.84TDD2070 pKa = 3.55VPTPLATDD2078 pKa = 3.2NCDD2081 pKa = 3.15IEE2083 pKa = 5.31ISATTDD2089 pKa = 3.1LVFPVTRR2096 pKa = 11.84QGSTMIRR2103 pKa = 11.84WKK2105 pKa = 11.03FEE2107 pKa = 4.03DD2108 pKa = 3.34SNANVTYY2115 pKa = 9.6QQQKK2119 pKa = 10.46IIIDD2123 pKa = 4.96DD2124 pKa = 3.44ITAPVPDD2131 pKa = 3.65IAEE2134 pKa = 4.38LEE2136 pKa = 4.42TVEE2139 pKa = 4.42VEE2141 pKa = 4.44CGVTAISAPTATDD2154 pKa = 2.93NCAGSVTGTTTDD2166 pKa = 3.45PLSYY2170 pKa = 10.45EE2171 pKa = 3.92EE2172 pKa = 4.07QGDD2175 pKa = 4.2YY2176 pKa = 11.0EE2177 pKa = 4.35ILWTFDD2183 pKa = 3.57DD2184 pKa = 4.83GNGNSTTQIQQVIVKK2199 pKa = 9.71DD2200 pKa = 3.47QTPPVALAKK2209 pKa = 10.52DD2210 pKa = 3.36ITVVLDD2216 pKa = 3.64PNDD2219 pKa = 3.39NVRR2222 pKa = 11.84INPEE2226 pKa = 4.49DD2227 pKa = 4.1IDD2229 pKa = 4.28DD2230 pKa = 4.25GSYY2233 pKa = 11.07DD2234 pKa = 3.65EE2235 pKa = 5.3CSEE2238 pKa = 4.08ISLSLDD2244 pKa = 2.86RR2245 pKa = 11.84DD2246 pKa = 4.01YY2247 pKa = 10.87FTSNGSYY2254 pKa = 10.23EE2255 pKa = 4.37VVLTVTDD2262 pKa = 3.83ALGNSSEE2269 pKa = 3.84ASARR2273 pKa = 11.84VIVVTNHH2280 pKa = 6.75PEE2282 pKa = 3.76TSNVHH2287 pKa = 4.77VVPTILKK2294 pKa = 7.92QSSMAKK2300 pKa = 9.59VIVPFRR2306 pKa = 11.84SRR2308 pKa = 11.84IIEE2311 pKa = 4.14VQVLEE2316 pKa = 4.45TEE2318 pKa = 4.22TNKK2321 pKa = 10.62YY2322 pKa = 10.31KK2323 pKa = 10.62IIRR2326 pKa = 11.84GNKK2329 pKa = 9.57LNEE2332 pKa = 3.89QQIDD2336 pKa = 3.33IAPFKK2341 pKa = 10.18GTLLVRR2347 pKa = 11.84IIDD2350 pKa = 3.47QDD2352 pKa = 3.65GRR2354 pKa = 11.84VHH2356 pKa = 6.86LKK2358 pKa = 10.68KK2359 pKa = 10.9LIALL2363 pKa = 4.36
Molecular weight: 250.43 kDa
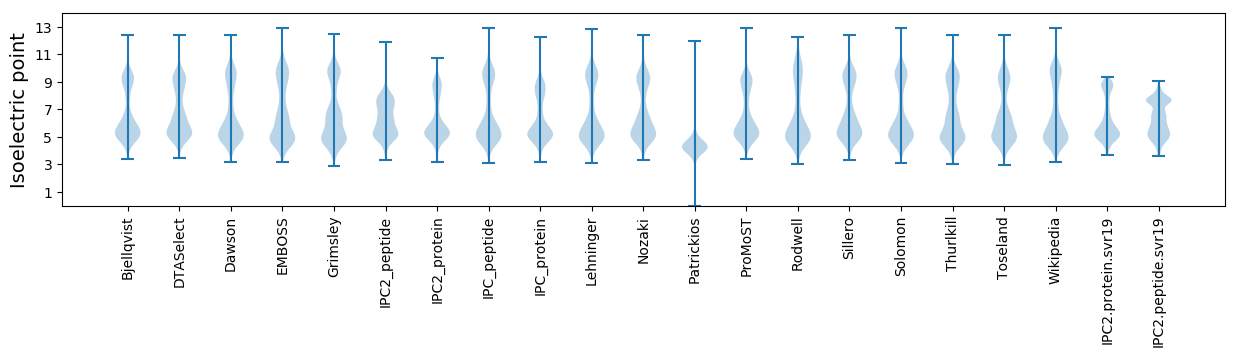
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3J3X5|A0A1L3J3X5_9FLAO Uncharacterized protein OS=Gramella salexigens OX=1913577 GN=LPB144_05140 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.73LSVSSEE47 pKa = 3.76NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
Molecular weight: 6.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
899170 |
44 |
5280 |
347.0 |
39.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.404 ± 0.049 | 0.694 ± 0.016 |
5.945 ± 0.063 | 7.408 ± 0.056 |
5.116 ± 0.044 | 6.619 ± 0.055 |
1.732 ± 0.027 | 7.72 ± 0.047 |
7.639 ± 0.077 | 9.274 ± 0.08 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.253 ± 0.031 | 5.806 ± 0.05 |
3.505 ± 0.031 | 3.207 ± 0.027 |
3.804 ± 0.047 | 6.655 ± 0.041 |
5.205 ± 0.1 | 6.027 ± 0.046 |
1.023 ± 0.018 | 3.965 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
