
Rubritalea squalenifaciens DSM 18772
Taxonomy: cellular organisms; Bacteria; PVC group; Verrucomicrobia; Verrucomicrobiae; Verrucomicrobiales; Rubritaleaceae; Rubritalea; Rubritalea squalenifaciens
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
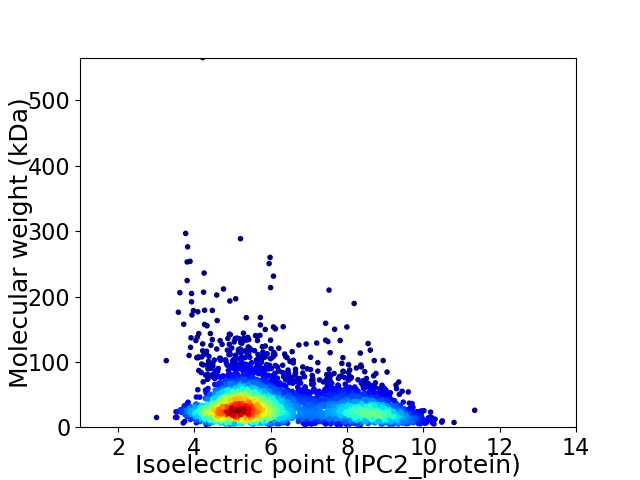
Virtual 2D-PAGE plot for 3805 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6ASZ7|A0A1M6ASZ7_9BACT N5-carboxyaminoimidazole ribonucleotide mutase OS=Rubritalea squalenifaciens DSM 18772 OX=1123071 GN=purE PE=3 SV=1
MM1 pKa = 7.27TPLIAFAQTLPDD13 pKa = 3.96SLVLNLDD20 pKa = 3.27TDD22 pKa = 4.5GNQTAEE28 pKa = 4.24TTVNLTKK35 pKa = 10.52RR36 pKa = 11.84SVRR39 pKa = 11.84APGVAYY45 pKa = 10.66YY46 pKa = 10.48RR47 pKa = 11.84FNGNSQTYY55 pKa = 8.14TLSPNDD61 pKa = 3.24IPQVRR66 pKa = 11.84TYY68 pKa = 10.41RR69 pKa = 11.84GTVAGDD75 pKa = 3.43PNVIVCATILPDD87 pKa = 3.6GTIKK91 pKa = 10.86ADD93 pKa = 3.91AFDD96 pKa = 3.87HH97 pKa = 6.19LKK99 pKa = 10.52GRR101 pKa = 11.84GYY103 pKa = 9.14VWQVSGYY110 pKa = 10.32DD111 pKa = 3.36VSASLNQGGAATANPTRR128 pKa = 11.84PEE130 pKa = 4.06GGMGEE135 pKa = 4.36LPMGVDD141 pKa = 3.16INYY144 pKa = 9.28HH145 pKa = 5.35RR146 pKa = 11.84FVNEE150 pKa = 3.92GASDD154 pKa = 4.28PDD156 pKa = 3.87LAWALTEE163 pKa = 4.46HH164 pKa = 6.54NFNIYY169 pKa = 10.65DD170 pKa = 3.72LMMGRR175 pKa = 11.84DD176 pKa = 3.22TGVAVSIATLVTRR189 pKa = 11.84EE190 pKa = 3.82NQEE193 pKa = 4.14YY194 pKa = 9.79YY195 pKa = 11.07VPTGSGNHH203 pKa = 5.73LNIIEE208 pKa = 4.75AEE210 pKa = 3.94WSDD213 pKa = 3.24AAKK216 pKa = 10.22PLRR219 pKa = 11.84NAAWEE224 pKa = 4.11QIFSYY229 pKa = 8.52KK230 pKa = 10.02TSVGLFGTGGYY241 pKa = 9.69AWQNRR246 pKa = 11.84IGKK249 pKa = 9.08NEE251 pKa = 4.23TINYY255 pKa = 7.07GQGAIGTQTLYY266 pKa = 10.95HH267 pKa = 6.81EE268 pKa = 5.02SGGHH272 pKa = 5.2NWNAHH277 pKa = 3.63HH278 pKa = 6.96WGYY281 pKa = 11.25GRR283 pKa = 11.84DD284 pKa = 3.5AMGSNRR290 pKa = 11.84PNHH293 pKa = 5.98GPYY296 pKa = 9.33NVEE299 pKa = 3.79RR300 pKa = 11.84VRR302 pKa = 11.84EE303 pKa = 3.93KK304 pKa = 10.88RR305 pKa = 11.84DD306 pKa = 3.13IKK308 pKa = 10.85IADD311 pKa = 3.74GTLLQPNGDD320 pKa = 4.26YY321 pKa = 10.53PSPAHH326 pKa = 6.78PYY328 pKa = 9.91SHH330 pKa = 7.57LDD332 pKa = 3.35LATTNVNQAVTISPLVNDD350 pKa = 3.89TDD352 pKa = 4.35FNGDD356 pKa = 3.75SISIVSFTATTANGGTVVQSGNDD379 pKa = 3.9LIYY382 pKa = 10.05TPPAGYY388 pKa = 9.71VGKK391 pKa = 10.22DD392 pKa = 3.05IIAYY396 pKa = 8.51TIEE399 pKa = 4.65DD400 pKa = 3.5NSGAGDD406 pKa = 3.81GGANLQAQDD415 pKa = 4.49LIFIEE420 pKa = 4.64VVNNGLTLKK429 pKa = 10.63FDD431 pKa = 4.41FNDD434 pKa = 3.06VSGASVSNSTGLGHH448 pKa = 6.62SAVLQSSNIEE458 pKa = 4.21SQLTTGPDD466 pKa = 3.49GYY468 pKa = 11.52ALVVPSGGLLVDD480 pKa = 4.37DD481 pKa = 4.87TNILPIPYY489 pKa = 8.86TGSTNNFYY497 pKa = 10.48PFDD500 pKa = 4.68SDD502 pKa = 4.91QMSQGNFFDD511 pKa = 5.7PMDD514 pKa = 4.67KK515 pKa = 10.69DD516 pKa = 3.68YY517 pKa = 11.42AICLWIKK524 pKa = 9.88MDD526 pKa = 5.08DD527 pKa = 4.15VITKK531 pKa = 10.46LDD533 pKa = 3.44ILDD536 pKa = 3.78KK537 pKa = 11.29YY538 pKa = 10.03KK539 pKa = 10.86AQEE542 pKa = 3.94RR543 pKa = 11.84STGFHH548 pKa = 6.4LFVEE552 pKa = 4.37NGVLKK557 pKa = 10.54ADD559 pKa = 3.83CGEE562 pKa = 4.01IEE564 pKa = 4.62GIYY567 pKa = 10.28NKK569 pKa = 10.36RR570 pKa = 11.84SITNGGAVTAGQWHH584 pKa = 6.29HH585 pKa = 6.12VALVFDD591 pKa = 4.38RR592 pKa = 11.84TADD595 pKa = 3.35QMKK598 pKa = 10.1IFLDD602 pKa = 4.73GAQTAQTALAAGSFIFHH619 pKa = 6.13GRR621 pKa = 11.84EE622 pKa = 3.43PLMIGSGSGGTMAVDD637 pKa = 3.96DD638 pKa = 4.2VRR640 pKa = 11.84LYY642 pKa = 9.08TQNLSSSDD650 pKa = 3.19LTGIMGEE657 pKa = 3.96ASNAVDD663 pKa = 4.54EE664 pKa = 4.64LALNNSLQTWWALDD678 pKa = 3.81DD679 pKa = 4.08NAGATASDD687 pKa = 3.37SSGNANAGALSGGAAWTTTAVDD709 pKa = 3.29GGGASFDD716 pKa = 3.95GVDD719 pKa = 5.02DD720 pKa = 5.29KK721 pKa = 11.64IKK723 pKa = 11.05LDD725 pKa = 3.86LAQSNVGAFTVAMWAKK741 pKa = 10.42SGADD745 pKa = 3.47GQGVYY750 pKa = 10.76SSVFSNRR757 pKa = 11.84TPNTGGTFQIDD768 pKa = 3.52MGNGFQYY775 pKa = 10.44RR776 pKa = 11.84GSQTSSFGAAPLDD789 pKa = 3.72QWVHH793 pKa = 5.7LAVVSDD799 pKa = 3.78GTDD802 pKa = 3.09TILYY806 pKa = 9.46YY807 pKa = 10.72NGEE810 pKa = 4.04IAKK813 pKa = 10.03VLSGVNDD820 pKa = 4.09DD821 pKa = 4.84LFNALVLGANRR832 pKa = 11.84NEE834 pKa = 3.86GDD836 pKa = 3.82FFQGEE841 pKa = 4.19IDD843 pKa = 3.8DD844 pKa = 3.85FCYY847 pKa = 9.74FDD849 pKa = 4.21RR850 pKa = 11.84ALNPSEE856 pKa = 3.6IQYY859 pKa = 10.44IKK861 pKa = 10.92SSLLKK866 pKa = 10.4NVAPSASDD874 pKa = 3.1DD875 pKa = 3.57TFAVVEE881 pKa = 4.11NASAGTSLGVVNASDD896 pKa = 4.18PNSGDD901 pKa = 3.72VVSHH905 pKa = 7.3AITGGNAAGLFAITAATGEE924 pKa = 4.31ITTTAPLDD932 pKa = 3.64YY933 pKa = 8.82EE934 pKa = 4.53TASSYY939 pKa = 11.54VLTVTVSDD947 pKa = 4.83DD948 pKa = 3.57GGLSDD953 pKa = 4.08TATITVNVTDD963 pKa = 4.03VANDD967 pKa = 3.69DD968 pKa = 3.93SDD970 pKa = 5.71ADD972 pKa = 3.82GLTDD976 pKa = 3.17EE977 pKa = 4.85WEE979 pKa = 4.17VANFGSVAATTGSADD994 pKa = 3.47SDD996 pKa = 3.65GDD998 pKa = 4.08GYY1000 pKa = 9.95TNAQEE1005 pKa = 4.19EE1006 pKa = 4.52ASATDD1011 pKa = 4.05PNNAASNPTTQVLAHH1026 pKa = 6.61WSLNEE1031 pKa = 3.79GAGLVATDD1039 pKa = 4.15SSVNGYY1045 pKa = 10.17DD1046 pKa = 3.52GQLTGGTAWTSSAVDD1061 pKa = 3.19AGGASFDD1068 pKa = 4.19GVDD1071 pKa = 4.66DD1072 pKa = 5.44RR1073 pKa = 11.84IDD1075 pKa = 3.49YY1076 pKa = 11.27ALTEE1080 pKa = 4.53TNLGAYY1086 pKa = 7.22TVAMWVKK1093 pKa = 10.38NGASGQSIYY1102 pKa = 11.38SSVFSNHH1109 pKa = 5.64TPNTSNTFQIDD1120 pKa = 3.18MGNGFNYY1127 pKa = 10.4RR1128 pKa = 11.84GTQTASFGAAPVGTWVHH1145 pKa = 6.12LAVVSDD1151 pKa = 3.87GTDD1154 pKa = 2.67TRR1156 pKa = 11.84LYY1158 pKa = 9.65YY1159 pKa = 10.81NGVLSQTLAGVNDD1172 pKa = 4.47DD1173 pKa = 4.83LFDD1176 pKa = 4.86TITLGTNRR1184 pKa = 11.84NEE1186 pKa = 4.11GNFFQGEE1193 pKa = 3.93VDD1195 pKa = 3.33EE1196 pKa = 5.65LYY1198 pKa = 11.07VYY1200 pKa = 10.68DD1201 pKa = 4.9AALNDD1206 pKa = 3.73EE1207 pKa = 5.71DD1208 pKa = 4.65ILALATPPAPNQPPVATNGSGSVAEE1233 pKa = 4.17NASVGTVVTTVVASDD1248 pKa = 4.36PDD1250 pKa = 3.76TGDD1253 pKa = 3.36TLSYY1257 pKa = 10.72SITAGNTGGAFAINSGTGEE1276 pKa = 4.13VTVAAGLDD1284 pKa = 3.79FEE1286 pKa = 6.33SISQYY1291 pKa = 10.74ILTVEE1296 pKa = 4.19VSDD1299 pKa = 5.48GSLTDD1304 pKa = 3.52TATVTIDD1311 pKa = 3.24VTNVNEE1317 pKa = 4.67APVANNGSGSVSEE1330 pKa = 4.47GASIGATVSTVSSSDD1345 pKa = 3.46PDD1347 pKa = 3.77TGDD1350 pKa = 3.14SATYY1354 pKa = 9.56TITAGNTGGAFVINSTSGAITTATGLDD1381 pKa = 3.96YY1382 pKa = 8.91EE1383 pKa = 4.72TTALYY1388 pKa = 10.95SLTVQVTDD1396 pKa = 4.22GGGLTDD1402 pKa = 3.64TATVTVNVTDD1412 pKa = 3.75VANDD1416 pKa = 3.69DD1417 pKa = 3.93SDD1419 pKa = 5.71ADD1421 pKa = 3.82GLTDD1425 pKa = 3.19EE1426 pKa = 5.01WEE1428 pKa = 4.13VSNFGSVAAQDD1439 pKa = 3.94GSGDD1443 pKa = 3.91ADD1445 pKa = 3.36SDD1447 pKa = 4.36GYY1449 pKa = 9.84TNAEE1453 pKa = 3.94EE1454 pKa = 4.32FAAGTDD1460 pKa = 3.6PSDD1463 pKa = 4.29PLSAPAPPDD1472 pKa = 3.71LVWTGAAGDD1481 pKa = 3.54GDD1483 pKa = 4.53FFNEE1487 pKa = 4.38ANWDD1491 pKa = 3.73SDD1493 pKa = 3.44ALTAGTQVPSADD1505 pKa = 3.36SVNVNQPLVASSLTVDD1521 pKa = 3.22SANFIGIGTVIPDD1534 pKa = 3.98GARR1537 pKa = 11.84IDD1539 pKa = 3.72ITNTTLTTSGSNGFNTTSSTLSEE1562 pKa = 4.32FNIGTGADD1570 pKa = 3.26VSFQFIANTTVSLAGDD1586 pKa = 3.43AVLEE1590 pKa = 4.16FRR1592 pKa = 11.84GGGNPVNGSTIDD1604 pKa = 3.69LAADD1608 pKa = 4.01FAGQVRR1614 pKa = 11.84FTSEE1618 pKa = 3.74TVASVLSEE1626 pKa = 4.14HH1627 pKa = 6.25MSKK1630 pKa = 8.48ITVAGQPAVNGVNAYY1645 pKa = 9.76VVDD1648 pKa = 4.14VNGVTVVSLEE1658 pKa = 4.15APNNAPVANDD1668 pKa = 3.24AAFTVAEE1675 pKa = 4.17NAGASATVGTVSVSDD1690 pKa = 4.29PDD1692 pKa = 5.53AGDD1695 pKa = 2.88SWTYY1699 pKa = 11.28AITAGNGGGEE1709 pKa = 4.05FAINSTTGEE1718 pKa = 4.07ITTTTSLDD1726 pKa = 3.53YY1727 pKa = 10.7EE1728 pKa = 4.66GASSYY1733 pKa = 11.51VLTVTVTDD1741 pKa = 3.81AGGLSDD1747 pKa = 3.74VATITVDD1754 pKa = 3.2VLNINEE1760 pKa = 4.16APVANSASGTVSEE1773 pKa = 4.79DD1774 pKa = 3.15VAVGVTVATVTSSDD1788 pKa = 3.4PDD1790 pKa = 3.79ANDD1793 pKa = 2.86TRR1795 pKa = 11.84TYY1797 pKa = 11.01AITAGNDD1804 pKa = 2.72GRR1806 pKa = 11.84FVINASTGEE1815 pKa = 4.03ITTVSALDD1823 pKa = 3.72YY1824 pKa = 7.19EE1825 pKa = 4.82TTNSYY1830 pKa = 11.33SLTVTVTDD1838 pKa = 4.16AGGLTATAMVAITVTDD1854 pKa = 3.8VVFEE1858 pKa = 5.34DD1859 pKa = 4.32YY1860 pKa = 11.13DD1861 pKa = 4.67SDD1863 pKa = 4.57SLDD1866 pKa = 4.49DD1867 pKa = 3.49NWEE1870 pKa = 3.76IANFGDD1876 pKa = 4.18TNTSDD1881 pKa = 3.39GTGDD1885 pKa = 3.6ADD1887 pKa = 5.38ADD1889 pKa = 3.82GLTDD1893 pKa = 3.78GEE1895 pKa = 4.57EE1896 pKa = 4.22YY1897 pKa = 10.9LAGSDD1902 pKa = 4.24PNSSDD1907 pKa = 3.15GDD1909 pKa = 3.43GDD1911 pKa = 3.89GFHH1914 pKa = 7.62DD1915 pKa = 3.71VLEE1918 pKa = 4.83INVGTDD1924 pKa = 3.03PMDD1927 pKa = 3.66ILDD1930 pKa = 4.86FPASEE1935 pKa = 4.21YY1936 pKa = 11.12DD1937 pKa = 5.19GLVLWWNLDD1946 pKa = 3.33EE1947 pKa = 4.66SAGASTVLDD1956 pKa = 3.45DD1957 pKa = 4.79TGNKK1961 pKa = 8.94MSGAVNGATLNGSEE1975 pKa = 4.52GSFDD1979 pKa = 3.8GSNDD1983 pKa = 4.39SIDD1986 pKa = 4.26LPLASGG1992 pKa = 3.65
MM1 pKa = 7.27TPLIAFAQTLPDD13 pKa = 3.96SLVLNLDD20 pKa = 3.27TDD22 pKa = 4.5GNQTAEE28 pKa = 4.24TTVNLTKK35 pKa = 10.52RR36 pKa = 11.84SVRR39 pKa = 11.84APGVAYY45 pKa = 10.66YY46 pKa = 10.48RR47 pKa = 11.84FNGNSQTYY55 pKa = 8.14TLSPNDD61 pKa = 3.24IPQVRR66 pKa = 11.84TYY68 pKa = 10.41RR69 pKa = 11.84GTVAGDD75 pKa = 3.43PNVIVCATILPDD87 pKa = 3.6GTIKK91 pKa = 10.86ADD93 pKa = 3.91AFDD96 pKa = 3.87HH97 pKa = 6.19LKK99 pKa = 10.52GRR101 pKa = 11.84GYY103 pKa = 9.14VWQVSGYY110 pKa = 10.32DD111 pKa = 3.36VSASLNQGGAATANPTRR128 pKa = 11.84PEE130 pKa = 4.06GGMGEE135 pKa = 4.36LPMGVDD141 pKa = 3.16INYY144 pKa = 9.28HH145 pKa = 5.35RR146 pKa = 11.84FVNEE150 pKa = 3.92GASDD154 pKa = 4.28PDD156 pKa = 3.87LAWALTEE163 pKa = 4.46HH164 pKa = 6.54NFNIYY169 pKa = 10.65DD170 pKa = 3.72LMMGRR175 pKa = 11.84DD176 pKa = 3.22TGVAVSIATLVTRR189 pKa = 11.84EE190 pKa = 3.82NQEE193 pKa = 4.14YY194 pKa = 9.79YY195 pKa = 11.07VPTGSGNHH203 pKa = 5.73LNIIEE208 pKa = 4.75AEE210 pKa = 3.94WSDD213 pKa = 3.24AAKK216 pKa = 10.22PLRR219 pKa = 11.84NAAWEE224 pKa = 4.11QIFSYY229 pKa = 8.52KK230 pKa = 10.02TSVGLFGTGGYY241 pKa = 9.69AWQNRR246 pKa = 11.84IGKK249 pKa = 9.08NEE251 pKa = 4.23TINYY255 pKa = 7.07GQGAIGTQTLYY266 pKa = 10.95HH267 pKa = 6.81EE268 pKa = 5.02SGGHH272 pKa = 5.2NWNAHH277 pKa = 3.63HH278 pKa = 6.96WGYY281 pKa = 11.25GRR283 pKa = 11.84DD284 pKa = 3.5AMGSNRR290 pKa = 11.84PNHH293 pKa = 5.98GPYY296 pKa = 9.33NVEE299 pKa = 3.79RR300 pKa = 11.84VRR302 pKa = 11.84EE303 pKa = 3.93KK304 pKa = 10.88RR305 pKa = 11.84DD306 pKa = 3.13IKK308 pKa = 10.85IADD311 pKa = 3.74GTLLQPNGDD320 pKa = 4.26YY321 pKa = 10.53PSPAHH326 pKa = 6.78PYY328 pKa = 9.91SHH330 pKa = 7.57LDD332 pKa = 3.35LATTNVNQAVTISPLVNDD350 pKa = 3.89TDD352 pKa = 4.35FNGDD356 pKa = 3.75SISIVSFTATTANGGTVVQSGNDD379 pKa = 3.9LIYY382 pKa = 10.05TPPAGYY388 pKa = 9.71VGKK391 pKa = 10.22DD392 pKa = 3.05IIAYY396 pKa = 8.51TIEE399 pKa = 4.65DD400 pKa = 3.5NSGAGDD406 pKa = 3.81GGANLQAQDD415 pKa = 4.49LIFIEE420 pKa = 4.64VVNNGLTLKK429 pKa = 10.63FDD431 pKa = 4.41FNDD434 pKa = 3.06VSGASVSNSTGLGHH448 pKa = 6.62SAVLQSSNIEE458 pKa = 4.21SQLTTGPDD466 pKa = 3.49GYY468 pKa = 11.52ALVVPSGGLLVDD480 pKa = 4.37DD481 pKa = 4.87TNILPIPYY489 pKa = 8.86TGSTNNFYY497 pKa = 10.48PFDD500 pKa = 4.68SDD502 pKa = 4.91QMSQGNFFDD511 pKa = 5.7PMDD514 pKa = 4.67KK515 pKa = 10.69DD516 pKa = 3.68YY517 pKa = 11.42AICLWIKK524 pKa = 9.88MDD526 pKa = 5.08DD527 pKa = 4.15VITKK531 pKa = 10.46LDD533 pKa = 3.44ILDD536 pKa = 3.78KK537 pKa = 11.29YY538 pKa = 10.03KK539 pKa = 10.86AQEE542 pKa = 3.94RR543 pKa = 11.84STGFHH548 pKa = 6.4LFVEE552 pKa = 4.37NGVLKK557 pKa = 10.54ADD559 pKa = 3.83CGEE562 pKa = 4.01IEE564 pKa = 4.62GIYY567 pKa = 10.28NKK569 pKa = 10.36RR570 pKa = 11.84SITNGGAVTAGQWHH584 pKa = 6.29HH585 pKa = 6.12VALVFDD591 pKa = 4.38RR592 pKa = 11.84TADD595 pKa = 3.35QMKK598 pKa = 10.1IFLDD602 pKa = 4.73GAQTAQTALAAGSFIFHH619 pKa = 6.13GRR621 pKa = 11.84EE622 pKa = 3.43PLMIGSGSGGTMAVDD637 pKa = 3.96DD638 pKa = 4.2VRR640 pKa = 11.84LYY642 pKa = 9.08TQNLSSSDD650 pKa = 3.19LTGIMGEE657 pKa = 3.96ASNAVDD663 pKa = 4.54EE664 pKa = 4.64LALNNSLQTWWALDD678 pKa = 3.81DD679 pKa = 4.08NAGATASDD687 pKa = 3.37SSGNANAGALSGGAAWTTTAVDD709 pKa = 3.29GGGASFDD716 pKa = 3.95GVDD719 pKa = 5.02DD720 pKa = 5.29KK721 pKa = 11.64IKK723 pKa = 11.05LDD725 pKa = 3.86LAQSNVGAFTVAMWAKK741 pKa = 10.42SGADD745 pKa = 3.47GQGVYY750 pKa = 10.76SSVFSNRR757 pKa = 11.84TPNTGGTFQIDD768 pKa = 3.52MGNGFQYY775 pKa = 10.44RR776 pKa = 11.84GSQTSSFGAAPLDD789 pKa = 3.72QWVHH793 pKa = 5.7LAVVSDD799 pKa = 3.78GTDD802 pKa = 3.09TILYY806 pKa = 9.46YY807 pKa = 10.72NGEE810 pKa = 4.04IAKK813 pKa = 10.03VLSGVNDD820 pKa = 4.09DD821 pKa = 4.84LFNALVLGANRR832 pKa = 11.84NEE834 pKa = 3.86GDD836 pKa = 3.82FFQGEE841 pKa = 4.19IDD843 pKa = 3.8DD844 pKa = 3.85FCYY847 pKa = 9.74FDD849 pKa = 4.21RR850 pKa = 11.84ALNPSEE856 pKa = 3.6IQYY859 pKa = 10.44IKK861 pKa = 10.92SSLLKK866 pKa = 10.4NVAPSASDD874 pKa = 3.1DD875 pKa = 3.57TFAVVEE881 pKa = 4.11NASAGTSLGVVNASDD896 pKa = 4.18PNSGDD901 pKa = 3.72VVSHH905 pKa = 7.3AITGGNAAGLFAITAATGEE924 pKa = 4.31ITTTAPLDD932 pKa = 3.64YY933 pKa = 8.82EE934 pKa = 4.53TASSYY939 pKa = 11.54VLTVTVSDD947 pKa = 4.83DD948 pKa = 3.57GGLSDD953 pKa = 4.08TATITVNVTDD963 pKa = 4.03VANDD967 pKa = 3.69DD968 pKa = 3.93SDD970 pKa = 5.71ADD972 pKa = 3.82GLTDD976 pKa = 3.17EE977 pKa = 4.85WEE979 pKa = 4.17VANFGSVAATTGSADD994 pKa = 3.47SDD996 pKa = 3.65GDD998 pKa = 4.08GYY1000 pKa = 9.95TNAQEE1005 pKa = 4.19EE1006 pKa = 4.52ASATDD1011 pKa = 4.05PNNAASNPTTQVLAHH1026 pKa = 6.61WSLNEE1031 pKa = 3.79GAGLVATDD1039 pKa = 4.15SSVNGYY1045 pKa = 10.17DD1046 pKa = 3.52GQLTGGTAWTSSAVDD1061 pKa = 3.19AGGASFDD1068 pKa = 4.19GVDD1071 pKa = 4.66DD1072 pKa = 5.44RR1073 pKa = 11.84IDD1075 pKa = 3.49YY1076 pKa = 11.27ALTEE1080 pKa = 4.53TNLGAYY1086 pKa = 7.22TVAMWVKK1093 pKa = 10.38NGASGQSIYY1102 pKa = 11.38SSVFSNHH1109 pKa = 5.64TPNTSNTFQIDD1120 pKa = 3.18MGNGFNYY1127 pKa = 10.4RR1128 pKa = 11.84GTQTASFGAAPVGTWVHH1145 pKa = 6.12LAVVSDD1151 pKa = 3.87GTDD1154 pKa = 2.67TRR1156 pKa = 11.84LYY1158 pKa = 9.65YY1159 pKa = 10.81NGVLSQTLAGVNDD1172 pKa = 4.47DD1173 pKa = 4.83LFDD1176 pKa = 4.86TITLGTNRR1184 pKa = 11.84NEE1186 pKa = 4.11GNFFQGEE1193 pKa = 3.93VDD1195 pKa = 3.33EE1196 pKa = 5.65LYY1198 pKa = 11.07VYY1200 pKa = 10.68DD1201 pKa = 4.9AALNDD1206 pKa = 3.73EE1207 pKa = 5.71DD1208 pKa = 4.65ILALATPPAPNQPPVATNGSGSVAEE1233 pKa = 4.17NASVGTVVTTVVASDD1248 pKa = 4.36PDD1250 pKa = 3.76TGDD1253 pKa = 3.36TLSYY1257 pKa = 10.72SITAGNTGGAFAINSGTGEE1276 pKa = 4.13VTVAAGLDD1284 pKa = 3.79FEE1286 pKa = 6.33SISQYY1291 pKa = 10.74ILTVEE1296 pKa = 4.19VSDD1299 pKa = 5.48GSLTDD1304 pKa = 3.52TATVTIDD1311 pKa = 3.24VTNVNEE1317 pKa = 4.67APVANNGSGSVSEE1330 pKa = 4.47GASIGATVSTVSSSDD1345 pKa = 3.46PDD1347 pKa = 3.77TGDD1350 pKa = 3.14SATYY1354 pKa = 9.56TITAGNTGGAFVINSTSGAITTATGLDD1381 pKa = 3.96YY1382 pKa = 8.91EE1383 pKa = 4.72TTALYY1388 pKa = 10.95SLTVQVTDD1396 pKa = 4.22GGGLTDD1402 pKa = 3.64TATVTVNVTDD1412 pKa = 3.75VANDD1416 pKa = 3.69DD1417 pKa = 3.93SDD1419 pKa = 5.71ADD1421 pKa = 3.82GLTDD1425 pKa = 3.19EE1426 pKa = 5.01WEE1428 pKa = 4.13VSNFGSVAAQDD1439 pKa = 3.94GSGDD1443 pKa = 3.91ADD1445 pKa = 3.36SDD1447 pKa = 4.36GYY1449 pKa = 9.84TNAEE1453 pKa = 3.94EE1454 pKa = 4.32FAAGTDD1460 pKa = 3.6PSDD1463 pKa = 4.29PLSAPAPPDD1472 pKa = 3.71LVWTGAAGDD1481 pKa = 3.54GDD1483 pKa = 4.53FFNEE1487 pKa = 4.38ANWDD1491 pKa = 3.73SDD1493 pKa = 3.44ALTAGTQVPSADD1505 pKa = 3.36SVNVNQPLVASSLTVDD1521 pKa = 3.22SANFIGIGTVIPDD1534 pKa = 3.98GARR1537 pKa = 11.84IDD1539 pKa = 3.72ITNTTLTTSGSNGFNTTSSTLSEE1562 pKa = 4.32FNIGTGADD1570 pKa = 3.26VSFQFIANTTVSLAGDD1586 pKa = 3.43AVLEE1590 pKa = 4.16FRR1592 pKa = 11.84GGGNPVNGSTIDD1604 pKa = 3.69LAADD1608 pKa = 4.01FAGQVRR1614 pKa = 11.84FTSEE1618 pKa = 3.74TVASVLSEE1626 pKa = 4.14HH1627 pKa = 6.25MSKK1630 pKa = 8.48ITVAGQPAVNGVNAYY1645 pKa = 9.76VVDD1648 pKa = 4.14VNGVTVVSLEE1658 pKa = 4.15APNNAPVANDD1668 pKa = 3.24AAFTVAEE1675 pKa = 4.17NAGASATVGTVSVSDD1690 pKa = 4.29PDD1692 pKa = 5.53AGDD1695 pKa = 2.88SWTYY1699 pKa = 11.28AITAGNGGGEE1709 pKa = 4.05FAINSTTGEE1718 pKa = 4.07ITTTTSLDD1726 pKa = 3.53YY1727 pKa = 10.7EE1728 pKa = 4.66GASSYY1733 pKa = 11.51VLTVTVTDD1741 pKa = 3.81AGGLSDD1747 pKa = 3.74VATITVDD1754 pKa = 3.2VLNINEE1760 pKa = 4.16APVANSASGTVSEE1773 pKa = 4.79DD1774 pKa = 3.15VAVGVTVATVTSSDD1788 pKa = 3.4PDD1790 pKa = 3.79ANDD1793 pKa = 2.86TRR1795 pKa = 11.84TYY1797 pKa = 11.01AITAGNDD1804 pKa = 2.72GRR1806 pKa = 11.84FVINASTGEE1815 pKa = 4.03ITTVSALDD1823 pKa = 3.72YY1824 pKa = 7.19EE1825 pKa = 4.82TTNSYY1830 pKa = 11.33SLTVTVTDD1838 pKa = 4.16AGGLTATAMVAITVTDD1854 pKa = 3.8VVFEE1858 pKa = 5.34DD1859 pKa = 4.32YY1860 pKa = 11.13DD1861 pKa = 4.67SDD1863 pKa = 4.57SLDD1866 pKa = 4.49DD1867 pKa = 3.49NWEE1870 pKa = 3.76IANFGDD1876 pKa = 4.18TNTSDD1881 pKa = 3.39GTGDD1885 pKa = 3.6ADD1887 pKa = 5.38ADD1889 pKa = 3.82GLTDD1893 pKa = 3.78GEE1895 pKa = 4.57EE1896 pKa = 4.22YY1897 pKa = 10.9LAGSDD1902 pKa = 4.24PNSSDD1907 pKa = 3.15GDD1909 pKa = 3.43GDD1911 pKa = 3.89GFHH1914 pKa = 7.62DD1915 pKa = 3.71VLEE1918 pKa = 4.83INVGTDD1924 pKa = 3.03PMDD1927 pKa = 3.66ILDD1930 pKa = 4.86FPASEE1935 pKa = 4.21YY1936 pKa = 11.12DD1937 pKa = 5.19GLVLWWNLDD1946 pKa = 3.33EE1947 pKa = 4.66SAGASTVLDD1956 pKa = 3.45DD1957 pKa = 4.79TGNKK1961 pKa = 8.94MSGAVNGATLNGSEE1975 pKa = 4.52GSFDD1979 pKa = 3.8GSNDD1983 pKa = 4.39SIDD1986 pKa = 4.26LPLASGG1992 pKa = 3.65
Molecular weight: 205.83 kDa
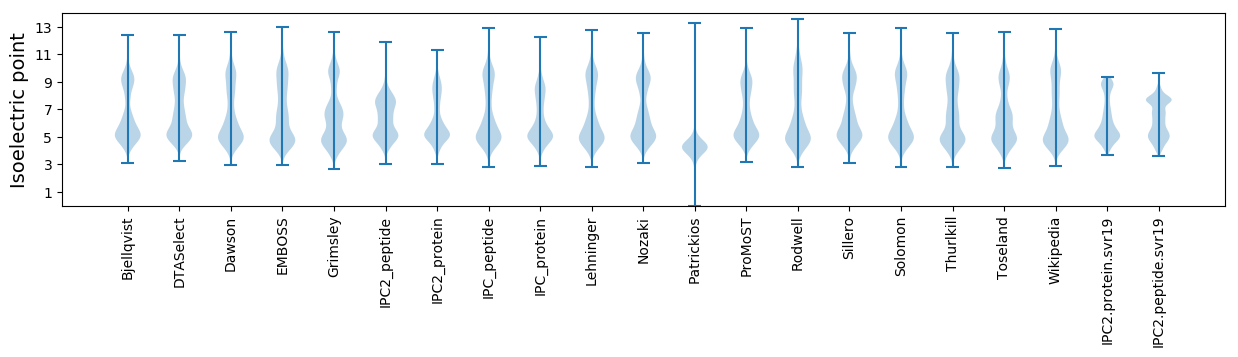
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6M0H0|A0A1M6M0H0_9BACT Thiol-disulfide interchange protein contains DsbC and DsbD domains OS=Rubritalea squalenifaciens DSM 18772 OX=1123071 GN=SAMN02745181_2559 PE=4 SV=1
MM1 pKa = 6.89STAHH5 pKa = 6.24EE6 pKa = 4.9KK7 pKa = 10.06IVHH10 pKa = 5.81AALRR14 pKa = 11.84DD15 pKa = 3.62ILEE18 pKa = 3.91QALRR22 pKa = 11.84RR23 pKa = 11.84LRR25 pKa = 11.84VVGGNDD31 pKa = 3.23LASKK35 pKa = 8.59VTVVWNSRR43 pKa = 11.84MRR45 pKa = 11.84STAGKK50 pKa = 9.59AHH52 pKa = 6.14WPEE55 pKa = 4.04AKK57 pKa = 10.38VEE59 pKa = 4.33LNPKK63 pKa = 10.13LIGISLAEE71 pKa = 4.19VKK73 pKa = 9.4RR74 pKa = 11.84TLLHH78 pKa = 6.27EE79 pKa = 4.42LAHH82 pKa = 6.23LLAYY86 pKa = 9.82HH87 pKa = 5.1RR88 pKa = 11.84HH89 pKa = 4.9KK90 pKa = 10.54RR91 pKa = 11.84RR92 pKa = 11.84RR93 pKa = 11.84IQPHH97 pKa = 4.21GRR99 pKa = 11.84EE100 pKa = 4.06WQQACADD107 pKa = 3.98LGIPGEE113 pKa = 4.52SVTHH117 pKa = 6.1NLALPSSRR125 pKa = 11.84QRR127 pKa = 11.84RR128 pKa = 11.84KK129 pKa = 8.05WRR131 pKa = 11.84YY132 pKa = 8.03VCPNCLQSIEE142 pKa = 4.38RR143 pKa = 11.84VRR145 pKa = 11.84KK146 pKa = 8.43MSASSACYY154 pKa = 9.68HH155 pKa = 6.76CCQNYY160 pKa = 10.52NGGKK164 pKa = 7.36YY165 pKa = 10.01HH166 pKa = 7.05SRR168 pKa = 11.84FRR170 pKa = 11.84LKK172 pKa = 10.74AEE174 pKa = 4.9RR175 pKa = 11.84IAA177 pKa = 4.61
MM1 pKa = 6.89STAHH5 pKa = 6.24EE6 pKa = 4.9KK7 pKa = 10.06IVHH10 pKa = 5.81AALRR14 pKa = 11.84DD15 pKa = 3.62ILEE18 pKa = 3.91QALRR22 pKa = 11.84RR23 pKa = 11.84LRR25 pKa = 11.84VVGGNDD31 pKa = 3.23LASKK35 pKa = 8.59VTVVWNSRR43 pKa = 11.84MRR45 pKa = 11.84STAGKK50 pKa = 9.59AHH52 pKa = 6.14WPEE55 pKa = 4.04AKK57 pKa = 10.38VEE59 pKa = 4.33LNPKK63 pKa = 10.13LIGISLAEE71 pKa = 4.19VKK73 pKa = 9.4RR74 pKa = 11.84TLLHH78 pKa = 6.27EE79 pKa = 4.42LAHH82 pKa = 6.23LLAYY86 pKa = 9.82HH87 pKa = 5.1RR88 pKa = 11.84HH89 pKa = 4.9KK90 pKa = 10.54RR91 pKa = 11.84RR92 pKa = 11.84RR93 pKa = 11.84IQPHH97 pKa = 4.21GRR99 pKa = 11.84EE100 pKa = 4.06WQQACADD107 pKa = 3.98LGIPGEE113 pKa = 4.52SVTHH117 pKa = 6.1NLALPSSRR125 pKa = 11.84QRR127 pKa = 11.84RR128 pKa = 11.84KK129 pKa = 8.05WRR131 pKa = 11.84YY132 pKa = 8.03VCPNCLQSIEE142 pKa = 4.38RR143 pKa = 11.84VRR145 pKa = 11.84KK146 pKa = 8.43MSASSACYY154 pKa = 9.68HH155 pKa = 6.76CCQNYY160 pKa = 10.52NGGKK164 pKa = 7.36YY165 pKa = 10.01HH166 pKa = 7.05SRR168 pKa = 11.84FRR170 pKa = 11.84LKK172 pKa = 10.74AEE174 pKa = 4.9RR175 pKa = 11.84IAA177 pKa = 4.61
Molecular weight: 20.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1291566 |
39 |
5242 |
339.4 |
37.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.554 ± 0.043 | 1.056 ± 0.014 |
5.861 ± 0.036 | 6.738 ± 0.048 |
3.876 ± 0.024 | 7.635 ± 0.052 |
2.281 ± 0.022 | 5.643 ± 0.028 |
5.661 ± 0.053 | 9.716 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.368 ± 0.022 | 3.766 ± 0.029 |
4.463 ± 0.027 | 3.638 ± 0.026 |
5.031 ± 0.038 | 6.83 ± 0.049 |
5.675 ± 0.047 | 6.614 ± 0.044 |
1.462 ± 0.016 | 3.132 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
