
Atractylodes mild mottle virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Caulimoviridae; Caulimovirus
Average proteome isoelectric point is 7.32
Get precalculated fractions of proteins
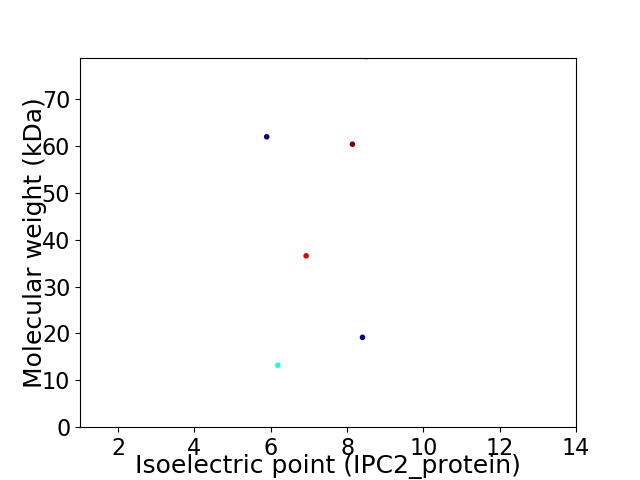
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
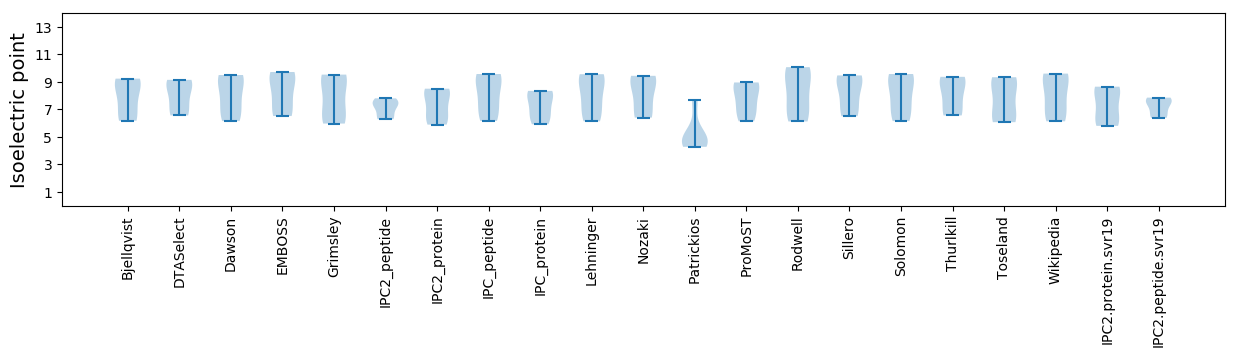
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M5K7P9|A0A0M5K7P9_9VIRU Protein 3 OS=Atractylodes mild mottle virus OX=1711685 PE=3 SV=1
MM1 pKa = 7.14TNYY4 pKa = 8.5MRR6 pKa = 11.84NVEE9 pKa = 4.15KK10 pKa = 10.66FFYY13 pKa = 10.72DD14 pKa = 3.18KK15 pKa = 11.38FEE17 pKa = 6.04DD18 pKa = 3.98DD19 pKa = 5.98LEE21 pKa = 4.57VTDD24 pKa = 4.44EE25 pKa = 4.33NYY27 pKa = 10.57HH28 pKa = 5.98ILNHH32 pKa = 5.8TLMLMDD38 pKa = 5.28MEE40 pKa = 5.79NEE42 pKa = 3.97DD43 pKa = 4.22DD44 pKa = 3.99ANSIIEE50 pKa = 4.51NDD52 pKa = 3.62PEE54 pKa = 4.15LKK56 pKa = 10.55KK57 pKa = 11.07FFGHH61 pKa = 7.4PDD63 pKa = 3.46PEE65 pKa = 4.3NLFVLGDD72 pKa = 3.88EE73 pKa = 5.31EE74 pKa = 4.54VSEE77 pKa = 4.41WNTEE81 pKa = 4.06SEE83 pKa = 4.64SEE85 pKa = 3.89FDD87 pKa = 4.98EE88 pKa = 5.88LIQDD92 pKa = 3.61EE93 pKa = 4.87PPRR96 pKa = 11.84KK97 pKa = 9.57KK98 pKa = 9.66PSEE101 pKa = 4.3RR102 pKa = 11.84IFPDD106 pKa = 2.94QQMIDD111 pKa = 3.74AYY113 pKa = 10.47AQSSYY118 pKa = 10.69TNPRR122 pKa = 11.84THH124 pKa = 6.41PTAYY128 pKa = 9.86QDD130 pKa = 3.17EE131 pKa = 5.01AYY133 pKa = 10.53SRR135 pKa = 11.84RR136 pKa = 11.84PNYY139 pKa = 9.97YY140 pKa = 9.36DD141 pKa = 3.68SKK143 pKa = 10.61DD144 pKa = 3.28IPSFYY149 pKa = 10.64DD150 pKa = 3.15KK151 pKa = 10.82KK152 pKa = 10.33FKK154 pKa = 10.7RR155 pKa = 11.84SAFVGSMSGNMSGSVQTQFGLSLNIDD181 pKa = 3.84CCGSAEE187 pKa = 4.28RR188 pKa = 11.84KK189 pKa = 9.4IKK191 pKa = 10.44LDD193 pKa = 3.08QWLNEE198 pKa = 4.2LNLMVQTNKK207 pKa = 10.39EE208 pKa = 4.06KK209 pKa = 10.66FDD211 pKa = 3.7TPKK214 pKa = 10.73KK215 pKa = 9.9VIILAEE221 pKa = 4.27HH222 pKa = 7.01KK223 pKa = 9.96STGVLQNFIKK233 pKa = 9.95RR234 pKa = 11.84TTWNVDD240 pKa = 3.19EE241 pKa = 5.41FSTSQMADD249 pKa = 2.59ILAEE253 pKa = 3.75AVYY256 pKa = 11.15ANFLGIDD263 pKa = 4.11FRR265 pKa = 11.84QSLVRR270 pKa = 11.84EE271 pKa = 4.25DD272 pKa = 3.34EE273 pKa = 4.27QIIKK277 pKa = 9.79KK278 pKa = 10.46AKK280 pKa = 9.3EE281 pKa = 4.11KK282 pKa = 9.11MSKK285 pKa = 7.83MTLCDD290 pKa = 2.93ICFLDD295 pKa = 5.04SFFCEE300 pKa = 4.4YY301 pKa = 10.71EE302 pKa = 4.33SAFHH306 pKa = 6.44SLKK309 pKa = 10.63EE310 pKa = 3.71RR311 pKa = 11.84DD312 pKa = 3.97DD313 pKa = 3.73FVKK316 pKa = 10.9FIEE319 pKa = 5.13LYY321 pKa = 9.21FAKK324 pKa = 10.31IPIVGSKK331 pKa = 10.1SLEE334 pKa = 3.9RR335 pKa = 11.84YY336 pKa = 9.41KK337 pKa = 10.78SEE339 pKa = 3.73KK340 pKa = 9.8TVVLEE345 pKa = 3.83TSIAYY350 pKa = 10.0ADD352 pKa = 4.43RR353 pKa = 11.84ITRR356 pKa = 11.84EE357 pKa = 4.26EE358 pKa = 3.72ISKK361 pKa = 10.09VCEE364 pKa = 3.69FSKK367 pKa = 9.76QQKK370 pKa = 9.37RR371 pKa = 11.84LKK373 pKa = 10.4KK374 pKa = 10.35FNKK377 pKa = 8.79NCCAGLVEE385 pKa = 4.48DD386 pKa = 4.24QNLEE390 pKa = 4.24FGCSISKK397 pKa = 10.46SNRR400 pKa = 11.84KK401 pKa = 9.21KK402 pKa = 10.74KK403 pKa = 10.74SFSKK407 pKa = 10.41KK408 pKa = 9.71KK409 pKa = 9.3KK410 pKa = 8.16KK411 pKa = 10.05KK412 pKa = 9.35YY413 pKa = 9.25YY414 pKa = 9.68KK415 pKa = 9.97KK416 pKa = 10.1SYY418 pKa = 8.3SKK420 pKa = 10.74KK421 pKa = 8.5RR422 pKa = 11.84VRR424 pKa = 11.84SKK426 pKa = 9.51FQPRR430 pKa = 11.84KK431 pKa = 9.42FFRR434 pKa = 11.84KK435 pKa = 9.58KK436 pKa = 10.25PPGKK440 pKa = 10.35EE441 pKa = 3.74KK442 pKa = 10.58FCPKK446 pKa = 10.08GKK448 pKa = 10.06KK449 pKa = 8.7SCRR452 pKa = 11.84CWICSEE458 pKa = 3.87QGHH461 pKa = 5.74YY462 pKa = 11.22ANEE465 pKa = 4.31CPNRR469 pKa = 11.84QAHH472 pKa = 4.91QKK474 pKa = 9.28QVNLFQEE481 pKa = 5.26AIQRR485 pKa = 11.84GLYY488 pKa = 9.55PIEE491 pKa = 4.54DD492 pKa = 4.25PYY494 pKa = 11.36EE495 pKa = 4.65GEE497 pKa = 3.76HH498 pKa = 6.13HH499 pKa = 6.54VYY501 pKa = 10.5EE502 pKa = 5.21FIVKK506 pKa = 9.91EE507 pKa = 4.2EE508 pKa = 4.27PPDD511 pKa = 3.74TDD513 pKa = 4.57SEE515 pKa = 4.79TTSSDD520 pKa = 3.02GSSSEE525 pKa = 4.33SSDD528 pKa = 3.85SEE530 pKa = 4.22
MM1 pKa = 7.14TNYY4 pKa = 8.5MRR6 pKa = 11.84NVEE9 pKa = 4.15KK10 pKa = 10.66FFYY13 pKa = 10.72DD14 pKa = 3.18KK15 pKa = 11.38FEE17 pKa = 6.04DD18 pKa = 3.98DD19 pKa = 5.98LEE21 pKa = 4.57VTDD24 pKa = 4.44EE25 pKa = 4.33NYY27 pKa = 10.57HH28 pKa = 5.98ILNHH32 pKa = 5.8TLMLMDD38 pKa = 5.28MEE40 pKa = 5.79NEE42 pKa = 3.97DD43 pKa = 4.22DD44 pKa = 3.99ANSIIEE50 pKa = 4.51NDD52 pKa = 3.62PEE54 pKa = 4.15LKK56 pKa = 10.55KK57 pKa = 11.07FFGHH61 pKa = 7.4PDD63 pKa = 3.46PEE65 pKa = 4.3NLFVLGDD72 pKa = 3.88EE73 pKa = 5.31EE74 pKa = 4.54VSEE77 pKa = 4.41WNTEE81 pKa = 4.06SEE83 pKa = 4.64SEE85 pKa = 3.89FDD87 pKa = 4.98EE88 pKa = 5.88LIQDD92 pKa = 3.61EE93 pKa = 4.87PPRR96 pKa = 11.84KK97 pKa = 9.57KK98 pKa = 9.66PSEE101 pKa = 4.3RR102 pKa = 11.84IFPDD106 pKa = 2.94QQMIDD111 pKa = 3.74AYY113 pKa = 10.47AQSSYY118 pKa = 10.69TNPRR122 pKa = 11.84THH124 pKa = 6.41PTAYY128 pKa = 9.86QDD130 pKa = 3.17EE131 pKa = 5.01AYY133 pKa = 10.53SRR135 pKa = 11.84RR136 pKa = 11.84PNYY139 pKa = 9.97YY140 pKa = 9.36DD141 pKa = 3.68SKK143 pKa = 10.61DD144 pKa = 3.28IPSFYY149 pKa = 10.64DD150 pKa = 3.15KK151 pKa = 10.82KK152 pKa = 10.33FKK154 pKa = 10.7RR155 pKa = 11.84SAFVGSMSGNMSGSVQTQFGLSLNIDD181 pKa = 3.84CCGSAEE187 pKa = 4.28RR188 pKa = 11.84KK189 pKa = 9.4IKK191 pKa = 10.44LDD193 pKa = 3.08QWLNEE198 pKa = 4.2LNLMVQTNKK207 pKa = 10.39EE208 pKa = 4.06KK209 pKa = 10.66FDD211 pKa = 3.7TPKK214 pKa = 10.73KK215 pKa = 9.9VIILAEE221 pKa = 4.27HH222 pKa = 7.01KK223 pKa = 9.96STGVLQNFIKK233 pKa = 9.95RR234 pKa = 11.84TTWNVDD240 pKa = 3.19EE241 pKa = 5.41FSTSQMADD249 pKa = 2.59ILAEE253 pKa = 3.75AVYY256 pKa = 11.15ANFLGIDD263 pKa = 4.11FRR265 pKa = 11.84QSLVRR270 pKa = 11.84EE271 pKa = 4.25DD272 pKa = 3.34EE273 pKa = 4.27QIIKK277 pKa = 9.79KK278 pKa = 10.46AKK280 pKa = 9.3EE281 pKa = 4.11KK282 pKa = 9.11MSKK285 pKa = 7.83MTLCDD290 pKa = 2.93ICFLDD295 pKa = 5.04SFFCEE300 pKa = 4.4YY301 pKa = 10.71EE302 pKa = 4.33SAFHH306 pKa = 6.44SLKK309 pKa = 10.63EE310 pKa = 3.71RR311 pKa = 11.84DD312 pKa = 3.97DD313 pKa = 3.73FVKK316 pKa = 10.9FIEE319 pKa = 5.13LYY321 pKa = 9.21FAKK324 pKa = 10.31IPIVGSKK331 pKa = 10.1SLEE334 pKa = 3.9RR335 pKa = 11.84YY336 pKa = 9.41KK337 pKa = 10.78SEE339 pKa = 3.73KK340 pKa = 9.8TVVLEE345 pKa = 3.83TSIAYY350 pKa = 10.0ADD352 pKa = 4.43RR353 pKa = 11.84ITRR356 pKa = 11.84EE357 pKa = 4.26EE358 pKa = 3.72ISKK361 pKa = 10.09VCEE364 pKa = 3.69FSKK367 pKa = 9.76QQKK370 pKa = 9.37RR371 pKa = 11.84LKK373 pKa = 10.4KK374 pKa = 10.35FNKK377 pKa = 8.79NCCAGLVEE385 pKa = 4.48DD386 pKa = 4.24QNLEE390 pKa = 4.24FGCSISKK397 pKa = 10.46SNRR400 pKa = 11.84KK401 pKa = 9.21KK402 pKa = 10.74KK403 pKa = 10.74SFSKK407 pKa = 10.41KK408 pKa = 9.71KK409 pKa = 9.3KK410 pKa = 8.16KK411 pKa = 10.05KK412 pKa = 9.35YY413 pKa = 9.25YY414 pKa = 9.68KK415 pKa = 9.97KK416 pKa = 10.1SYY418 pKa = 8.3SKK420 pKa = 10.74KK421 pKa = 8.5RR422 pKa = 11.84VRR424 pKa = 11.84SKK426 pKa = 9.51FQPRR430 pKa = 11.84KK431 pKa = 9.42FFRR434 pKa = 11.84KK435 pKa = 9.58KK436 pKa = 10.25PPGKK440 pKa = 10.35EE441 pKa = 3.74KK442 pKa = 10.58FCPKK446 pKa = 10.08GKK448 pKa = 10.06KK449 pKa = 8.7SCRR452 pKa = 11.84CWICSEE458 pKa = 3.87QGHH461 pKa = 5.74YY462 pKa = 11.22ANEE465 pKa = 4.31CPNRR469 pKa = 11.84QAHH472 pKa = 4.91QKK474 pKa = 9.28QVNLFQEE481 pKa = 5.26AIQRR485 pKa = 11.84GLYY488 pKa = 9.55PIEE491 pKa = 4.54DD492 pKa = 4.25PYY494 pKa = 11.36EE495 pKa = 4.65GEE497 pKa = 3.76HH498 pKa = 6.13HH499 pKa = 6.54VYY501 pKa = 10.5EE502 pKa = 5.21FIVKK506 pKa = 9.91EE507 pKa = 4.2EE508 pKa = 4.27PPDD511 pKa = 3.74TDD513 pKa = 4.57SEE515 pKa = 4.79TTSSDD520 pKa = 3.02GSSSEE525 pKa = 4.33SSDD528 pKa = 3.85SEE530 pKa = 4.22
Molecular weight: 61.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M4J5R0|A0A0M4J5R0_9VIRU Aphid transmission protein OS=Atractylodes mild mottle virus OX=1711685 PE=3 SV=1
MM1 pKa = 8.12DD2 pKa = 6.41LLQKK6 pKa = 10.51VQIQNNHH13 pKa = 4.72QQLSLNATNTSSIYY27 pKa = 10.28ISGKK31 pKa = 10.55LKK33 pKa = 10.18FQGYY37 pKa = 8.64KK38 pKa = 10.12QLDD41 pKa = 3.48LHH43 pKa = 6.69CFVDD47 pKa = 4.6TGASMCVASKK57 pKa = 10.62HH58 pKa = 5.91VIPEE62 pKa = 4.12EE63 pKa = 3.6HH64 pKa = 6.89WEE66 pKa = 4.03NSSRR70 pKa = 11.84QLLVKK75 pKa = 8.86TANGMITINKK85 pKa = 6.96VCKK88 pKa = 10.63NIDD91 pKa = 2.98IMIADD96 pKa = 5.09TIFHH100 pKa = 6.99IPTIYY105 pKa = 9.76QQEE108 pKa = 4.01IGIDD112 pKa = 3.56LLLGNNFLLLYY123 pKa = 10.84GPFTQYY129 pKa = 9.33TDD131 pKa = 4.16RR132 pKa = 11.84IILFKK137 pKa = 10.7EE138 pKa = 3.8NQPVIIGKK146 pKa = 8.56VSKK149 pKa = 10.41AYY151 pKa = 10.01LHH153 pKa = 6.41GMPGYY158 pKa = 10.15LDD160 pKa = 3.69SMKK163 pKa = 10.56KK164 pKa = 10.2NSRR167 pKa = 11.84NPVPPPINITSNKK180 pKa = 9.5IEE182 pKa = 4.17DD183 pKa = 3.66SKK185 pKa = 11.28RR186 pKa = 11.84FSVLKK191 pKa = 10.49LQLLEE196 pKa = 4.16RR197 pKa = 11.84GRR199 pKa = 11.84EE200 pKa = 3.93IINRR204 pKa = 11.84RR205 pKa = 11.84LFLTQQDD212 pKa = 3.69QLSTIEE218 pKa = 4.19KK219 pKa = 10.38LLDD222 pKa = 3.69EE223 pKa = 4.86VCSEE227 pKa = 4.56NPIDD231 pKa = 3.6PHH233 pKa = 8.2KK234 pKa = 10.46SKK236 pKa = 10.58KK237 pKa = 9.29WMTASIKK244 pKa = 10.88LKK246 pKa = 10.75DD247 pKa = 4.07PNTLVQVKK255 pKa = 9.22PMQYY259 pKa = 9.91CPEE262 pKa = 4.08DD263 pKa = 3.61RR264 pKa = 11.84KK265 pKa = 10.86EE266 pKa = 3.7FAVQIKK272 pKa = 9.94EE273 pKa = 4.04LLDD276 pKa = 3.66LKK278 pKa = 10.75IIIPSKK284 pKa = 10.12SPHH287 pKa = 6.66RR288 pKa = 11.84SPAFLVEE295 pKa = 4.16NEE297 pKa = 3.65AEE299 pKa = 3.89RR300 pKa = 11.84RR301 pKa = 11.84RR302 pKa = 11.84GKK304 pKa = 10.54KK305 pKa = 10.14RR306 pKa = 11.84MVVNYY311 pKa = 9.91KK312 pKa = 10.22KK313 pKa = 10.82LNEE316 pKa = 4.03VTLGDD321 pKa = 4.29SHH323 pKa = 6.93NLPNKK328 pKa = 10.49DD329 pKa = 3.68EE330 pKa = 4.84LLTLIRR336 pKa = 11.84GKK338 pKa = 9.05TIYY341 pKa = 10.73SSFDD345 pKa = 3.5CKK347 pKa = 10.96SGFWQVLLDD356 pKa = 4.39QEE358 pKa = 4.77SQLLTAFTCPTGHH371 pKa = 5.73YY372 pKa = 7.52QWKK375 pKa = 9.56VVPFGLKK382 pKa = 8.89QAPSIFQRR390 pKa = 11.84HH391 pKa = 4.9MNNAFRR397 pKa = 11.84EE398 pKa = 4.29FEE400 pKa = 4.33EE401 pKa = 5.12FCCVYY406 pKa = 10.75VDD408 pKa = 5.62DD409 pKa = 5.43ILVFSKK415 pKa = 11.31NEE417 pKa = 3.86TEE419 pKa = 4.12HH420 pKa = 6.43RR421 pKa = 11.84KK422 pKa = 9.31HH423 pKa = 6.69VIAILEE429 pKa = 4.02QCKK432 pKa = 10.29NLGIILSKK440 pKa = 10.75KK441 pKa = 8.95KK442 pKa = 10.55AHH444 pKa = 6.62LFKK447 pKa = 10.79TKK449 pKa = 10.36INFLGLEE456 pKa = 4.11IDD458 pKa = 3.8QGSHH462 pKa = 6.36KK463 pKa = 9.39PQNHH467 pKa = 5.81ILEE470 pKa = 5.46HH471 pKa = 5.17IHH473 pKa = 6.66KK474 pKa = 10.3FPDD477 pKa = 3.12RR478 pKa = 11.84LEE480 pKa = 4.65DD481 pKa = 3.39KK482 pKa = 10.78KK483 pKa = 11.01QLQRR487 pKa = 11.84FLGILTYY494 pKa = 10.95ASDD497 pKa = 4.78YY498 pKa = 10.58IPKK501 pKa = 9.73LAQIRR506 pKa = 11.84KK507 pKa = 8.53PFQAKK512 pKa = 9.74LKK514 pKa = 10.76KK515 pKa = 10.28DD516 pKa = 3.96VTWSWTDD523 pKa = 2.68SDD525 pKa = 4.03TSLMIKK531 pKa = 9.72IKK533 pKa = 10.46KK534 pKa = 8.69GLKK537 pKa = 9.88SFPTLYY543 pKa = 10.43HH544 pKa = 6.54PKK546 pKa = 10.55EE547 pKa = 4.0EE548 pKa = 4.67DD549 pKa = 3.31NLIIEE554 pKa = 5.12CDD556 pKa = 3.45ASDD559 pKa = 4.06SFWGGILKK567 pKa = 10.6AKK569 pKa = 9.84TIDD572 pKa = 3.37EE573 pKa = 4.42DD574 pKa = 3.61KK575 pKa = 11.01EE576 pKa = 4.37YY577 pKa = 10.45ICRR580 pKa = 11.84YY581 pKa = 7.54TSGSFKK587 pKa = 10.4QAEE590 pKa = 4.42LNYY593 pKa = 10.1HH594 pKa = 5.34SNEE597 pKa = 3.92KK598 pKa = 10.48EE599 pKa = 3.42ILAAMNTIKK608 pKa = 10.64KK609 pKa = 9.93FSGYY613 pKa = 7.92LTPVKK618 pKa = 10.31FLIRR622 pKa = 11.84TDD624 pKa = 3.25NKK626 pKa = 10.66NFTFFLNTNHH636 pKa = 7.12KK637 pKa = 10.45GDD639 pKa = 3.9YY640 pKa = 9.4KK641 pKa = 10.28QGRR644 pKa = 11.84LIRR647 pKa = 11.84WQMWFSRR654 pKa = 11.84YY655 pKa = 9.88SFTVEE660 pKa = 4.0HH661 pKa = 6.67LPGNKK666 pKa = 9.3NVFADD671 pKa = 4.28FLTRR675 pKa = 11.84EE676 pKa = 4.33FNNN679 pKa = 3.45
MM1 pKa = 8.12DD2 pKa = 6.41LLQKK6 pKa = 10.51VQIQNNHH13 pKa = 4.72QQLSLNATNTSSIYY27 pKa = 10.28ISGKK31 pKa = 10.55LKK33 pKa = 10.18FQGYY37 pKa = 8.64KK38 pKa = 10.12QLDD41 pKa = 3.48LHH43 pKa = 6.69CFVDD47 pKa = 4.6TGASMCVASKK57 pKa = 10.62HH58 pKa = 5.91VIPEE62 pKa = 4.12EE63 pKa = 3.6HH64 pKa = 6.89WEE66 pKa = 4.03NSSRR70 pKa = 11.84QLLVKK75 pKa = 8.86TANGMITINKK85 pKa = 6.96VCKK88 pKa = 10.63NIDD91 pKa = 2.98IMIADD96 pKa = 5.09TIFHH100 pKa = 6.99IPTIYY105 pKa = 9.76QQEE108 pKa = 4.01IGIDD112 pKa = 3.56LLLGNNFLLLYY123 pKa = 10.84GPFTQYY129 pKa = 9.33TDD131 pKa = 4.16RR132 pKa = 11.84IILFKK137 pKa = 10.7EE138 pKa = 3.8NQPVIIGKK146 pKa = 8.56VSKK149 pKa = 10.41AYY151 pKa = 10.01LHH153 pKa = 6.41GMPGYY158 pKa = 10.15LDD160 pKa = 3.69SMKK163 pKa = 10.56KK164 pKa = 10.2NSRR167 pKa = 11.84NPVPPPINITSNKK180 pKa = 9.5IEE182 pKa = 4.17DD183 pKa = 3.66SKK185 pKa = 11.28RR186 pKa = 11.84FSVLKK191 pKa = 10.49LQLLEE196 pKa = 4.16RR197 pKa = 11.84GRR199 pKa = 11.84EE200 pKa = 3.93IINRR204 pKa = 11.84RR205 pKa = 11.84LFLTQQDD212 pKa = 3.69QLSTIEE218 pKa = 4.19KK219 pKa = 10.38LLDD222 pKa = 3.69EE223 pKa = 4.86VCSEE227 pKa = 4.56NPIDD231 pKa = 3.6PHH233 pKa = 8.2KK234 pKa = 10.46SKK236 pKa = 10.58KK237 pKa = 9.29WMTASIKK244 pKa = 10.88LKK246 pKa = 10.75DD247 pKa = 4.07PNTLVQVKK255 pKa = 9.22PMQYY259 pKa = 9.91CPEE262 pKa = 4.08DD263 pKa = 3.61RR264 pKa = 11.84KK265 pKa = 10.86EE266 pKa = 3.7FAVQIKK272 pKa = 9.94EE273 pKa = 4.04LLDD276 pKa = 3.66LKK278 pKa = 10.75IIIPSKK284 pKa = 10.12SPHH287 pKa = 6.66RR288 pKa = 11.84SPAFLVEE295 pKa = 4.16NEE297 pKa = 3.65AEE299 pKa = 3.89RR300 pKa = 11.84RR301 pKa = 11.84RR302 pKa = 11.84GKK304 pKa = 10.54KK305 pKa = 10.14RR306 pKa = 11.84MVVNYY311 pKa = 9.91KK312 pKa = 10.22KK313 pKa = 10.82LNEE316 pKa = 4.03VTLGDD321 pKa = 4.29SHH323 pKa = 6.93NLPNKK328 pKa = 10.49DD329 pKa = 3.68EE330 pKa = 4.84LLTLIRR336 pKa = 11.84GKK338 pKa = 9.05TIYY341 pKa = 10.73SSFDD345 pKa = 3.5CKK347 pKa = 10.96SGFWQVLLDD356 pKa = 4.39QEE358 pKa = 4.77SQLLTAFTCPTGHH371 pKa = 5.73YY372 pKa = 7.52QWKK375 pKa = 9.56VVPFGLKK382 pKa = 8.89QAPSIFQRR390 pKa = 11.84HH391 pKa = 4.9MNNAFRR397 pKa = 11.84EE398 pKa = 4.29FEE400 pKa = 4.33EE401 pKa = 5.12FCCVYY406 pKa = 10.75VDD408 pKa = 5.62DD409 pKa = 5.43ILVFSKK415 pKa = 11.31NEE417 pKa = 3.86TEE419 pKa = 4.12HH420 pKa = 6.43RR421 pKa = 11.84KK422 pKa = 9.31HH423 pKa = 6.69VIAILEE429 pKa = 4.02QCKK432 pKa = 10.29NLGIILSKK440 pKa = 10.75KK441 pKa = 8.95KK442 pKa = 10.55AHH444 pKa = 6.62LFKK447 pKa = 10.79TKK449 pKa = 10.36INFLGLEE456 pKa = 4.11IDD458 pKa = 3.8QGSHH462 pKa = 6.36KK463 pKa = 9.39PQNHH467 pKa = 5.81ILEE470 pKa = 5.46HH471 pKa = 5.17IHH473 pKa = 6.66KK474 pKa = 10.3FPDD477 pKa = 3.12RR478 pKa = 11.84LEE480 pKa = 4.65DD481 pKa = 3.39KK482 pKa = 10.78KK483 pKa = 11.01QLQRR487 pKa = 11.84FLGILTYY494 pKa = 10.95ASDD497 pKa = 4.78YY498 pKa = 10.58IPKK501 pKa = 9.73LAQIRR506 pKa = 11.84KK507 pKa = 8.53PFQAKK512 pKa = 9.74LKK514 pKa = 10.76KK515 pKa = 10.28DD516 pKa = 3.96VTWSWTDD523 pKa = 2.68SDD525 pKa = 4.03TSLMIKK531 pKa = 9.72IKK533 pKa = 10.46KK534 pKa = 8.69GLKK537 pKa = 9.88SFPTLYY543 pKa = 10.43HH544 pKa = 6.54PKK546 pKa = 10.55EE547 pKa = 4.0EE548 pKa = 4.67DD549 pKa = 3.31NLIIEE554 pKa = 5.12CDD556 pKa = 3.45ASDD559 pKa = 4.06SFWGGILKK567 pKa = 10.6AKK569 pKa = 9.84TIDD572 pKa = 3.37EE573 pKa = 4.42DD574 pKa = 3.61KK575 pKa = 11.01EE576 pKa = 4.37YY577 pKa = 10.45ICRR580 pKa = 11.84YY581 pKa = 7.54TSGSFKK587 pKa = 10.4QAEE590 pKa = 4.42LNYY593 pKa = 10.1HH594 pKa = 5.34SNEE597 pKa = 3.92KK598 pKa = 10.48EE599 pKa = 3.42ILAAMNTIKK608 pKa = 10.64KK609 pKa = 9.93FSGYY613 pKa = 7.92LTPVKK618 pKa = 10.31FLIRR622 pKa = 11.84TDD624 pKa = 3.25NKK626 pKa = 10.66NFTFFLNTNHH636 pKa = 7.12KK637 pKa = 10.45GDD639 pKa = 3.9YY640 pKa = 9.4KK641 pKa = 10.28QGRR644 pKa = 11.84LIRR647 pKa = 11.84WQMWFSRR654 pKa = 11.84YY655 pKa = 9.88SFTVEE660 pKa = 4.0HH661 pKa = 6.67LPGNKK666 pKa = 9.3NVFADD671 pKa = 4.28FLTRR675 pKa = 11.84EE676 pKa = 4.33FNNN679 pKa = 3.45
Molecular weight: 78.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2358 |
120 |
679 |
393.0 |
45.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.411 ± 0.455 | 1.739 ± 0.238 |
5.895 ± 0.356 | 7.591 ± 0.741 |
5.047 ± 0.408 | 4.241 ± 0.248 |
2.417 ± 0.342 | 6.955 ± 0.539 |
11.323 ± 0.62 | 8.567 ± 0.78 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.696 ± 0.212 | 5.768 ± 0.579 |
4.368 ± 0.5 | 4.283 ± 0.309 |
3.562 ± 0.332 | 8.482 ± 0.649 |
4.665 ± 0.235 | 5.089 ± 0.477 |
0.721 ± 0.187 | 3.181 ± 0.24 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
