
Nocardia terpenica
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Nocardiaceae; Nocardia
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
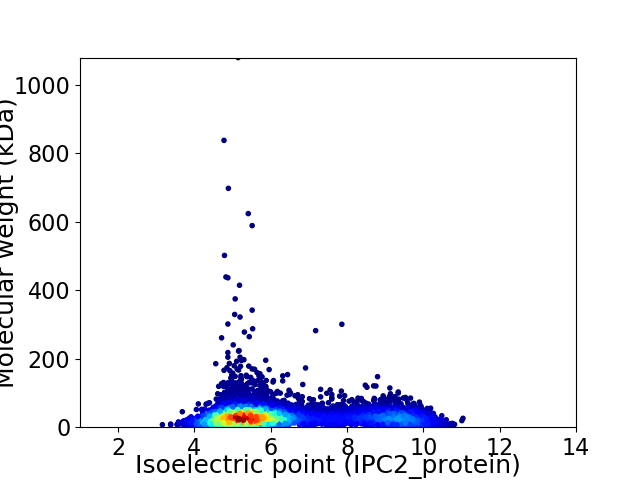
Virtual 2D-PAGE plot for 8083 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A164K9H6|A0A164K9H6_9NOCA SARP family transcriptional regulator OS=Nocardia terpenica OX=455432 GN=AWN90_38665 PE=3 SV=1
MM1 pKa = 7.82ADD3 pKa = 3.36LASVYY8 pKa = 10.85DD9 pKa = 4.36EE10 pKa = 4.9AAQDD14 pKa = 3.69TPEE17 pKa = 3.89WAEE20 pKa = 3.43WDD22 pKa = 3.61DD23 pKa = 3.63PRR25 pKa = 11.84RR26 pKa = 11.84SFMQIDD32 pKa = 3.68TLFDD36 pKa = 3.32EE37 pKa = 5.12TLPALPVADD46 pKa = 4.62DD47 pKa = 3.67AALGEE52 pKa = 4.6TVPPRR57 pKa = 11.84PQAPRR62 pKa = 11.84YY63 pKa = 8.89SPAMLEE69 pKa = 4.16WVEE72 pKa = 4.04YY73 pKa = 10.51AAFAMFPTRR82 pKa = 11.84KK83 pKa = 9.79ALADD87 pKa = 3.5NTDD90 pKa = 3.28AADD93 pKa = 3.5QFVCYY98 pKa = 10.28LIEE101 pKa = 4.09YY102 pKa = 9.28LVRR105 pKa = 11.84NAGGLRR111 pKa = 11.84FNVPGNGSPVYY122 pKa = 10.49DD123 pKa = 3.34GFGPSVCYY131 pKa = 10.49NYY133 pKa = 11.06ASDD136 pKa = 3.59VDD138 pKa = 4.16NPVDD142 pKa = 3.42MLLTIGEE149 pKa = 4.38HH150 pKa = 6.59GEE152 pKa = 4.18GFADD156 pKa = 5.44DD157 pKa = 4.64IITRR161 pKa = 11.84AEE163 pKa = 4.38DD164 pKa = 3.68YY165 pKa = 11.26ADD167 pKa = 3.56SATT170 pKa = 3.69
MM1 pKa = 7.82ADD3 pKa = 3.36LASVYY8 pKa = 10.85DD9 pKa = 4.36EE10 pKa = 4.9AAQDD14 pKa = 3.69TPEE17 pKa = 3.89WAEE20 pKa = 3.43WDD22 pKa = 3.61DD23 pKa = 3.63PRR25 pKa = 11.84RR26 pKa = 11.84SFMQIDD32 pKa = 3.68TLFDD36 pKa = 3.32EE37 pKa = 5.12TLPALPVADD46 pKa = 4.62DD47 pKa = 3.67AALGEE52 pKa = 4.6TVPPRR57 pKa = 11.84PQAPRR62 pKa = 11.84YY63 pKa = 8.89SPAMLEE69 pKa = 4.16WVEE72 pKa = 4.04YY73 pKa = 10.51AAFAMFPTRR82 pKa = 11.84KK83 pKa = 9.79ALADD87 pKa = 3.5NTDD90 pKa = 3.28AADD93 pKa = 3.5QFVCYY98 pKa = 10.28LIEE101 pKa = 4.09YY102 pKa = 9.28LVRR105 pKa = 11.84NAGGLRR111 pKa = 11.84FNVPGNGSPVYY122 pKa = 10.49DD123 pKa = 3.34GFGPSVCYY131 pKa = 10.49NYY133 pKa = 11.06ASDD136 pKa = 3.59VDD138 pKa = 4.16NPVDD142 pKa = 3.42MLLTIGEE149 pKa = 4.38HH150 pKa = 6.59GEE152 pKa = 4.18GFADD156 pKa = 5.44DD157 pKa = 4.64IITRR161 pKa = 11.84AEE163 pKa = 4.38DD164 pKa = 3.68YY165 pKa = 11.26ADD167 pKa = 3.56SATT170 pKa = 3.69
Molecular weight: 18.69 kDa
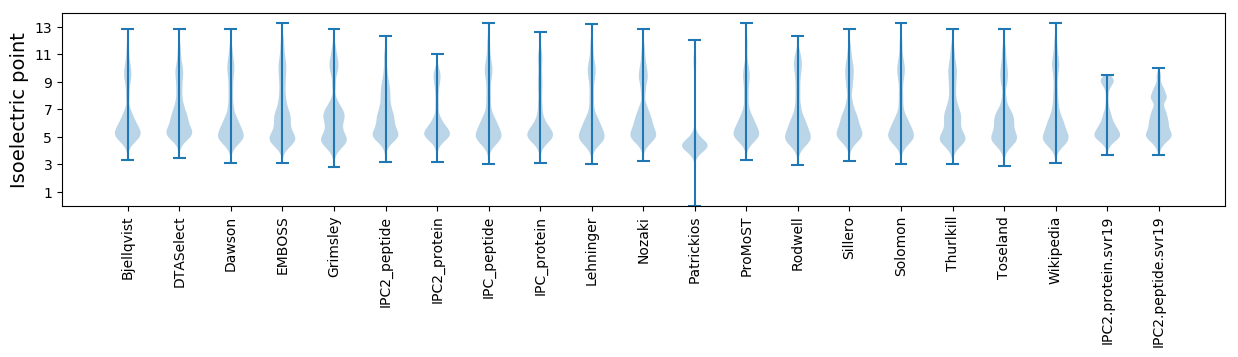
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A164NWR9|A0A164NWR9_9NOCA Bifunctional 3'-5' exonuclease/DNA polymerase OS=Nocardia terpenica OX=455432 GN=AWN90_22565 PE=4 SV=1
MM1 pKa = 7.82PLLPLGLAAPAGRR14 pKa = 11.84ATGTAEE20 pKa = 4.18GTRR23 pKa = 11.84GTAATAAATRR33 pKa = 11.84TATAATGTTPEE44 pKa = 3.83ATATGRR50 pKa = 11.84GSARR54 pKa = 11.84TGAGTAATAAVVTTAAATGTTAAAGRR80 pKa = 11.84ATRR83 pKa = 11.84TARR86 pKa = 11.84TATRR90 pKa = 11.84RR91 pKa = 11.84RR92 pKa = 11.84ARR94 pKa = 11.84TRR96 pKa = 11.84RR97 pKa = 11.84ALRR100 pKa = 11.84HH101 pKa = 4.52GTRR104 pKa = 11.84VRR106 pKa = 11.84PRR108 pKa = 11.84GHH110 pKa = 6.83RR111 pKa = 11.84AGARR115 pKa = 11.84PRR117 pKa = 11.84TTRR120 pKa = 11.84TRR122 pKa = 11.84AATLGGRR129 pKa = 11.84RR130 pKa = 11.84TLPARR135 pKa = 11.84GRR137 pKa = 11.84TLSTRR142 pKa = 11.84SRR144 pKa = 11.84ALPARR149 pKa = 11.84GGTRR153 pKa = 11.84AALGRR158 pKa = 11.84TRR160 pKa = 11.84ARR162 pKa = 11.84ARR164 pKa = 11.84LRR166 pKa = 11.84PRR168 pKa = 11.84RR169 pKa = 11.84GTLGRR174 pKa = 11.84GAEE177 pKa = 4.16GVVADD182 pKa = 4.41ARR184 pKa = 11.84GLRR187 pKa = 11.84TRR189 pKa = 11.84LRR191 pKa = 11.84AGPARR196 pKa = 11.84TRR198 pKa = 11.84ALMRR202 pKa = 11.84GSAALVLATGTRR214 pKa = 11.84GARR217 pKa = 11.84SRR219 pKa = 11.84PLVALLLTVVVLLLRR234 pKa = 11.84RR235 pKa = 11.84GRR237 pKa = 11.84GPGVAPDD244 pKa = 3.73AGFADD249 pKa = 3.85GRR251 pKa = 11.84AWLL254 pKa = 4.09
MM1 pKa = 7.82PLLPLGLAAPAGRR14 pKa = 11.84ATGTAEE20 pKa = 4.18GTRR23 pKa = 11.84GTAATAAATRR33 pKa = 11.84TATAATGTTPEE44 pKa = 3.83ATATGRR50 pKa = 11.84GSARR54 pKa = 11.84TGAGTAATAAVVTTAAATGTTAAAGRR80 pKa = 11.84ATRR83 pKa = 11.84TARR86 pKa = 11.84TATRR90 pKa = 11.84RR91 pKa = 11.84RR92 pKa = 11.84ARR94 pKa = 11.84TRR96 pKa = 11.84RR97 pKa = 11.84ALRR100 pKa = 11.84HH101 pKa = 4.52GTRR104 pKa = 11.84VRR106 pKa = 11.84PRR108 pKa = 11.84GHH110 pKa = 6.83RR111 pKa = 11.84AGARR115 pKa = 11.84PRR117 pKa = 11.84TTRR120 pKa = 11.84TRR122 pKa = 11.84AATLGGRR129 pKa = 11.84RR130 pKa = 11.84TLPARR135 pKa = 11.84GRR137 pKa = 11.84TLSTRR142 pKa = 11.84SRR144 pKa = 11.84ALPARR149 pKa = 11.84GGTRR153 pKa = 11.84AALGRR158 pKa = 11.84TRR160 pKa = 11.84ARR162 pKa = 11.84ARR164 pKa = 11.84LRR166 pKa = 11.84PRR168 pKa = 11.84RR169 pKa = 11.84GTLGRR174 pKa = 11.84GAEE177 pKa = 4.16GVVADD182 pKa = 4.41ARR184 pKa = 11.84GLRR187 pKa = 11.84TRR189 pKa = 11.84LRR191 pKa = 11.84AGPARR196 pKa = 11.84TRR198 pKa = 11.84ALMRR202 pKa = 11.84GSAALVLATGTRR214 pKa = 11.84GARR217 pKa = 11.84SRR219 pKa = 11.84PLVALLLTVVVLLLRR234 pKa = 11.84RR235 pKa = 11.84GRR237 pKa = 11.84GPGVAPDD244 pKa = 3.73AGFADD249 pKa = 3.85GRR251 pKa = 11.84AWLL254 pKa = 4.09
Molecular weight: 25.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2507138 |
37 |
10051 |
310.2 |
33.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.455 ± 0.046 | 0.811 ± 0.008 |
6.236 ± 0.022 | 5.206 ± 0.027 |
2.886 ± 0.015 | 8.851 ± 0.029 |
2.346 ± 0.013 | 4.078 ± 0.017 |
1.8 ± 0.02 | 10.215 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.864 ± 0.013 | 1.943 ± 0.015 |
5.993 ± 0.025 | 2.811 ± 0.02 |
8.207 ± 0.033 | 4.999 ± 0.018 |
6.215 ± 0.021 | 8.423 ± 0.026 |
1.539 ± 0.01 | 2.122 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
