
Clostridiaceae bacterium
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; unclassified Clostridiaceae
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
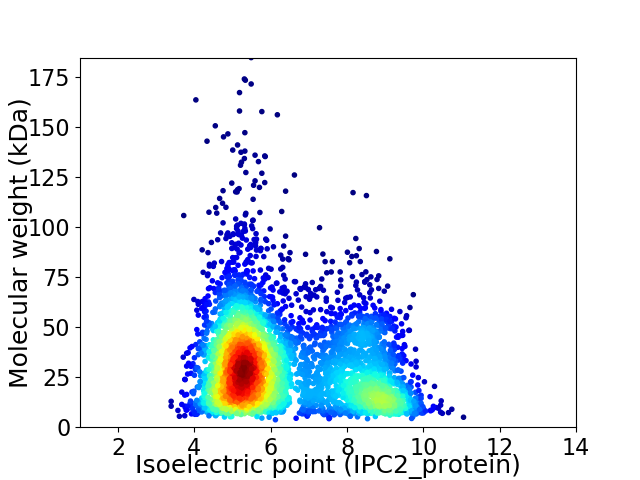
Virtual 2D-PAGE plot for 4379 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6L9H7C2|A0A6L9H7C2_9CLOT HlyC/CorC family transporter OS=Clostridiaceae bacterium OX=1898204 GN=D3Z50_03740 PE=3 SV=1
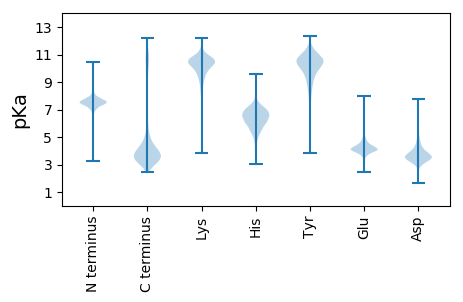
MM1 pKa = 7.57KK2 pKa = 10.56KK3 pKa = 8.92MLSVAMAGMLALSLTACGSSEE24 pKa = 4.63KK25 pKa = 10.06PAEE28 pKa = 4.29TQAPAPTEE36 pKa = 3.98AVKK39 pKa = 10.21TEE41 pKa = 3.93AAVEE45 pKa = 4.11EE46 pKa = 4.61AVTEE50 pKa = 4.1AVKK53 pKa = 10.7EE54 pKa = 4.09DD55 pKa = 3.49AASEE59 pKa = 4.06GGYY62 pKa = 9.13TIGFSPYY69 pKa = 9.64TLTNEE74 pKa = 4.01YY75 pKa = 8.52FTAVQTGVQAACDD88 pKa = 3.66EE89 pKa = 5.11LGCEE93 pKa = 4.75MISFDD98 pKa = 4.7PQNDD102 pKa = 3.54PTKK105 pKa = 10.25QASQIEE111 pKa = 4.3DD112 pKa = 3.74MIASGIDD119 pKa = 3.09ALVYY123 pKa = 10.22IPYY126 pKa = 10.1DD127 pKa = 3.11SAGAHH132 pKa = 5.32TVLQTCRR139 pKa = 11.84DD140 pKa = 3.55AGIKK144 pKa = 9.44VINVDD149 pKa = 3.77NVITEE154 pKa = 4.2DD155 pKa = 4.61DD156 pKa = 4.07YY157 pKa = 12.08EE158 pKa = 4.38LVDD161 pKa = 6.25GIIASDD167 pKa = 3.71NTQLGYY173 pKa = 11.19LSGQWVAANHH183 pKa = 6.83PDD185 pKa = 3.36GANILIVHH193 pKa = 6.7LQTAEE198 pKa = 3.84SCIINVEE205 pKa = 4.44GFWSGIKK212 pKa = 10.59DD213 pKa = 3.59NVSNADD219 pKa = 3.24AFVEE223 pKa = 4.38VQVVEE228 pKa = 4.45GEE230 pKa = 4.45GATDD234 pKa = 3.1AAFNVVSDD242 pKa = 4.45ALQAHH247 pKa = 7.32DD248 pKa = 6.0DD249 pKa = 3.71IDD251 pKa = 4.35VIYY254 pKa = 10.25CINDD258 pKa = 3.44TSALGAVQAVEE269 pKa = 4.24EE270 pKa = 4.25AGKK273 pKa = 10.58AGSIDD278 pKa = 3.47ILGKK282 pKa = 10.48DD283 pKa = 3.44GAPIGKK289 pKa = 8.83HH290 pKa = 5.47AIKK293 pKa = 10.4DD294 pKa = 3.58GTMVQSSAQRR304 pKa = 11.84PTYY307 pKa = 8.39MGYY310 pKa = 9.03MGVQNAVKK318 pKa = 9.91ALKK321 pKa = 10.81GEE323 pKa = 3.98EE324 pKa = 4.21FEE326 pKa = 4.51FNIAIEE332 pKa = 4.53SYY334 pKa = 10.93SIDD337 pKa = 3.37ASNIDD342 pKa = 5.67DD343 pKa = 4.37YY344 pKa = 12.14DD345 pKa = 5.04LDD347 pKa = 3.68AWDD350 pKa = 5.08ALDD353 pKa = 3.56
MM1 pKa = 7.57KK2 pKa = 10.56KK3 pKa = 8.92MLSVAMAGMLALSLTACGSSEE24 pKa = 4.63KK25 pKa = 10.06PAEE28 pKa = 4.29TQAPAPTEE36 pKa = 3.98AVKK39 pKa = 10.21TEE41 pKa = 3.93AAVEE45 pKa = 4.11EE46 pKa = 4.61AVTEE50 pKa = 4.1AVKK53 pKa = 10.7EE54 pKa = 4.09DD55 pKa = 3.49AASEE59 pKa = 4.06GGYY62 pKa = 9.13TIGFSPYY69 pKa = 9.64TLTNEE74 pKa = 4.01YY75 pKa = 8.52FTAVQTGVQAACDD88 pKa = 3.66EE89 pKa = 5.11LGCEE93 pKa = 4.75MISFDD98 pKa = 4.7PQNDD102 pKa = 3.54PTKK105 pKa = 10.25QASQIEE111 pKa = 4.3DD112 pKa = 3.74MIASGIDD119 pKa = 3.09ALVYY123 pKa = 10.22IPYY126 pKa = 10.1DD127 pKa = 3.11SAGAHH132 pKa = 5.32TVLQTCRR139 pKa = 11.84DD140 pKa = 3.55AGIKK144 pKa = 9.44VINVDD149 pKa = 3.77NVITEE154 pKa = 4.2DD155 pKa = 4.61DD156 pKa = 4.07YY157 pKa = 12.08EE158 pKa = 4.38LVDD161 pKa = 6.25GIIASDD167 pKa = 3.71NTQLGYY173 pKa = 11.19LSGQWVAANHH183 pKa = 6.83PDD185 pKa = 3.36GANILIVHH193 pKa = 6.7LQTAEE198 pKa = 3.84SCIINVEE205 pKa = 4.44GFWSGIKK212 pKa = 10.59DD213 pKa = 3.59NVSNADD219 pKa = 3.24AFVEE223 pKa = 4.38VQVVEE228 pKa = 4.45GEE230 pKa = 4.45GATDD234 pKa = 3.1AAFNVVSDD242 pKa = 4.45ALQAHH247 pKa = 7.32DD248 pKa = 6.0DD249 pKa = 3.71IDD251 pKa = 4.35VIYY254 pKa = 10.25CINDD258 pKa = 3.44TSALGAVQAVEE269 pKa = 4.24EE270 pKa = 4.25AGKK273 pKa = 10.58AGSIDD278 pKa = 3.47ILGKK282 pKa = 10.48DD283 pKa = 3.44GAPIGKK289 pKa = 8.83HH290 pKa = 5.47AIKK293 pKa = 10.4DD294 pKa = 3.58GTMVQSSAQRR304 pKa = 11.84PTYY307 pKa = 8.39MGYY310 pKa = 9.03MGVQNAVKK318 pKa = 9.91ALKK321 pKa = 10.81GEE323 pKa = 3.98EE324 pKa = 4.21FEE326 pKa = 4.51FNIAIEE332 pKa = 4.53SYY334 pKa = 10.93SIDD337 pKa = 3.37ASNIDD342 pKa = 5.67DD343 pKa = 4.37YY344 pKa = 12.14DD345 pKa = 5.04LDD347 pKa = 3.68AWDD350 pKa = 5.08ALDD353 pKa = 3.56
Molecular weight: 37.26 kDa
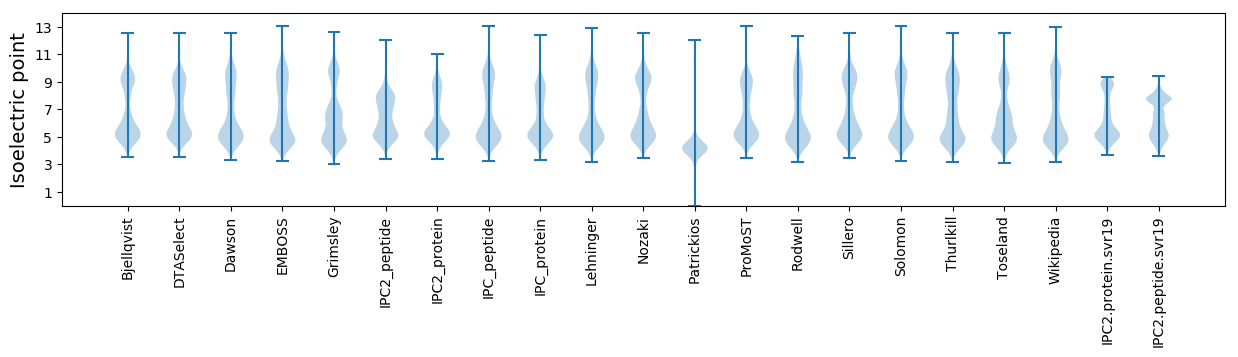
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6L9HCR9|A0A6L9HCR9_9CLOT ATP-binding protein OS=Clostridiaceae bacterium OX=1898204 GN=D3Z50_05840 PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 8.56MTYY5 pKa = 8.56QPKK8 pKa = 9.32KK9 pKa = 7.54RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.81VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.6KK2 pKa = 8.56MTYY5 pKa = 8.56QPKK8 pKa = 9.32KK9 pKa = 7.54RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.81VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 4.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1312029 |
35 |
1611 |
299.6 |
33.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.643 ± 0.042 | 1.581 ± 0.016 |
5.384 ± 0.03 | 7.688 ± 0.043 |
4.119 ± 0.027 | 7.872 ± 0.04 |
1.687 ± 0.016 | 6.523 ± 0.033 |
5.816 ± 0.035 | 9.16 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.077 ± 0.017 | 3.664 ± 0.027 |
3.572 ± 0.022 | 3.174 ± 0.021 |
5.912 ± 0.04 | 5.797 ± 0.031 |
4.875 ± 0.026 | 6.438 ± 0.029 |
1.033 ± 0.011 | 3.985 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
