
Cyclobacterium lianum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cyclobacteriaceae; Cyclobacterium
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
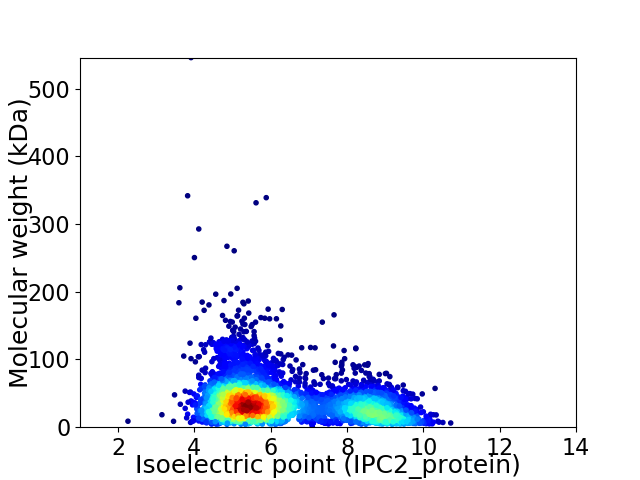
Virtual 2D-PAGE plot for 4679 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M7NUB9|A0A1M7NUB9_9BACT Glucose/arabinose dehydrogenase beta-propeller fold OS=Cyclobacterium lianum OX=388280 GN=SAMN04488057_10678 PE=4 SV=1
MM1 pKa = 6.68MTYY4 pKa = 10.3KK5 pKa = 10.49HH6 pKa = 5.89MRR8 pKa = 11.84IEE10 pKa = 4.09NVTYY14 pKa = 10.43RR15 pKa = 11.84GVLTLLFGSLCIISCEE31 pKa = 4.13TADD34 pKa = 4.13TGTGMDD40 pKa = 4.41DD41 pKa = 3.48PGLPGSTGSTFACTEE56 pKa = 3.97FSYY59 pKa = 11.53SDD61 pKa = 3.06TLYY64 pKa = 10.87FINEE68 pKa = 3.86QTEE71 pKa = 4.05QLINPQTEE79 pKa = 3.97LAGTYY84 pKa = 9.59SASPEE89 pKa = 4.06GLDD92 pKa = 3.99IDD94 pKa = 4.55PLSGAIDD101 pKa = 3.42INASEE106 pKa = 4.63TGLKK110 pKa = 9.49YY111 pKa = 10.49QISFSPDD118 pKa = 2.64NGAPSCPVFLTIGGINYY135 pKa = 9.47LDD137 pKa = 3.3QVYY140 pKa = 10.54VLDD143 pKa = 3.9QGEE146 pKa = 4.35TQAIPVYY153 pKa = 10.65NGVPSAAIPCEE164 pKa = 4.68DD165 pKa = 5.62DD166 pKa = 6.3DD167 pKa = 7.7DD168 pKa = 7.6DD169 pKa = 7.69DD170 pKa = 7.66DD171 pKa = 7.68DD172 pKa = 7.65DD173 pKa = 7.65DD174 pKa = 7.65DD175 pKa = 7.65DD176 pKa = 7.65DD177 pKa = 7.65DD178 pKa = 7.65DD179 pKa = 7.61DD180 pKa = 7.59DD181 pKa = 7.58DD182 pKa = 7.79DD183 pKa = 5.57DD184 pKa = 5.25DD185 pKa = 5.55CRR187 pKa = 11.84FDD189 pKa = 4.99ADD191 pKa = 4.97GPDD194 pKa = 3.75GQNLADD200 pKa = 4.42AGVAINASTGTIDD213 pKa = 6.33LEE215 pKa = 4.44TTLNNGALGSSPANGTFSDD234 pKa = 4.68FSLYY238 pKa = 10.45YY239 pKa = 10.4RR240 pKa = 11.84LNDD243 pKa = 3.74PSMGALNRR251 pKa = 11.84IGLRR255 pKa = 11.84VYY257 pKa = 10.85YY258 pKa = 10.44FEE260 pKa = 4.28TTADD264 pKa = 3.74VPQSLLDD271 pKa = 4.38EE272 pKa = 4.92IEE274 pKa = 4.22TKK276 pKa = 10.6RR277 pKa = 11.84NQILGTQATGQHH289 pKa = 6.0LRR291 pKa = 11.84TSNDD295 pKa = 2.98NPRR298 pKa = 11.84VRR300 pKa = 11.84PRR302 pKa = 11.84PPYY305 pKa = 9.87LVIVSRR311 pKa = 11.84LRR313 pKa = 3.41
MM1 pKa = 6.68MTYY4 pKa = 10.3KK5 pKa = 10.49HH6 pKa = 5.89MRR8 pKa = 11.84IEE10 pKa = 4.09NVTYY14 pKa = 10.43RR15 pKa = 11.84GVLTLLFGSLCIISCEE31 pKa = 4.13TADD34 pKa = 4.13TGTGMDD40 pKa = 4.41DD41 pKa = 3.48PGLPGSTGSTFACTEE56 pKa = 3.97FSYY59 pKa = 11.53SDD61 pKa = 3.06TLYY64 pKa = 10.87FINEE68 pKa = 3.86QTEE71 pKa = 4.05QLINPQTEE79 pKa = 3.97LAGTYY84 pKa = 9.59SASPEE89 pKa = 4.06GLDD92 pKa = 3.99IDD94 pKa = 4.55PLSGAIDD101 pKa = 3.42INASEE106 pKa = 4.63TGLKK110 pKa = 9.49YY111 pKa = 10.49QISFSPDD118 pKa = 2.64NGAPSCPVFLTIGGINYY135 pKa = 9.47LDD137 pKa = 3.3QVYY140 pKa = 10.54VLDD143 pKa = 3.9QGEE146 pKa = 4.35TQAIPVYY153 pKa = 10.65NGVPSAAIPCEE164 pKa = 4.68DD165 pKa = 5.62DD166 pKa = 6.3DD167 pKa = 7.7DD168 pKa = 7.6DD169 pKa = 7.69DD170 pKa = 7.66DD171 pKa = 7.68DD172 pKa = 7.65DD173 pKa = 7.65DD174 pKa = 7.65DD175 pKa = 7.65DD176 pKa = 7.65DD177 pKa = 7.65DD178 pKa = 7.65DD179 pKa = 7.61DD180 pKa = 7.59DD181 pKa = 7.58DD182 pKa = 7.79DD183 pKa = 5.57DD184 pKa = 5.25DD185 pKa = 5.55CRR187 pKa = 11.84FDD189 pKa = 4.99ADD191 pKa = 4.97GPDD194 pKa = 3.75GQNLADD200 pKa = 4.42AGVAINASTGTIDD213 pKa = 6.33LEE215 pKa = 4.44TTLNNGALGSSPANGTFSDD234 pKa = 4.68FSLYY238 pKa = 10.45YY239 pKa = 10.4RR240 pKa = 11.84LNDD243 pKa = 3.74PSMGALNRR251 pKa = 11.84IGLRR255 pKa = 11.84VYY257 pKa = 10.85YY258 pKa = 10.44FEE260 pKa = 4.28TTADD264 pKa = 3.74VPQSLLDD271 pKa = 4.38EE272 pKa = 4.92IEE274 pKa = 4.22TKK276 pKa = 10.6RR277 pKa = 11.84NQILGTQATGQHH289 pKa = 6.0LRR291 pKa = 11.84TSNDD295 pKa = 2.98NPRR298 pKa = 11.84VRR300 pKa = 11.84PRR302 pKa = 11.84PPYY305 pKa = 9.87LVIVSRR311 pKa = 11.84LRR313 pKa = 3.41
Molecular weight: 33.89 kDa
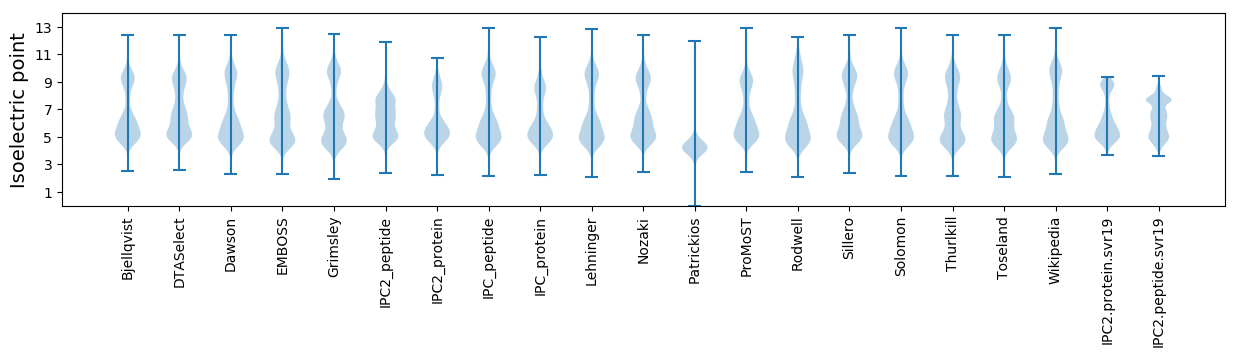
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M7PV89|A0A1M7PV89_9BACT RND family efflux transporter MFP subunit OS=Cyclobacterium lianum OX=388280 GN=SAMN04488057_1114 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.18RR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VLKK32 pKa = 10.53ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.17GRR39 pKa = 11.84HH40 pKa = 5.24KK41 pKa = 10.87LSVSSEE47 pKa = 3.97KK48 pKa = 9.91TLKK51 pKa = 10.51KK52 pKa = 10.72
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.18RR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VLKK32 pKa = 10.53ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.17GRR39 pKa = 11.84HH40 pKa = 5.24KK41 pKa = 10.87LSVSSEE47 pKa = 3.97KK48 pKa = 9.91TLKK51 pKa = 10.51KK52 pKa = 10.72
Molecular weight: 6.22 kDa
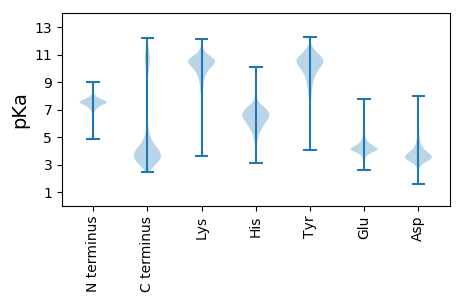
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1677476 |
10 |
5137 |
358.5 |
40.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.116 ± 0.03 | 0.7 ± 0.011 |
5.572 ± 0.027 | 6.805 ± 0.03 |
5.084 ± 0.028 | 7.391 ± 0.032 |
1.975 ± 0.018 | 6.735 ± 0.029 |
5.897 ± 0.043 | 9.985 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.453 ± 0.017 | 4.94 ± 0.036 |
4.217 ± 0.023 | 3.798 ± 0.02 |
4.723 ± 0.023 | 6.437 ± 0.024 |
4.907 ± 0.023 | 6.127 ± 0.026 |
1.35 ± 0.014 | 3.787 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
