
Drosophila x virus (isolate Chung/1996) (DXV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Birnaviridae; Entomobirnavirus; Drosophila X virus
Average proteome isoelectric point is 7.42
Get precalculated fractions of proteins
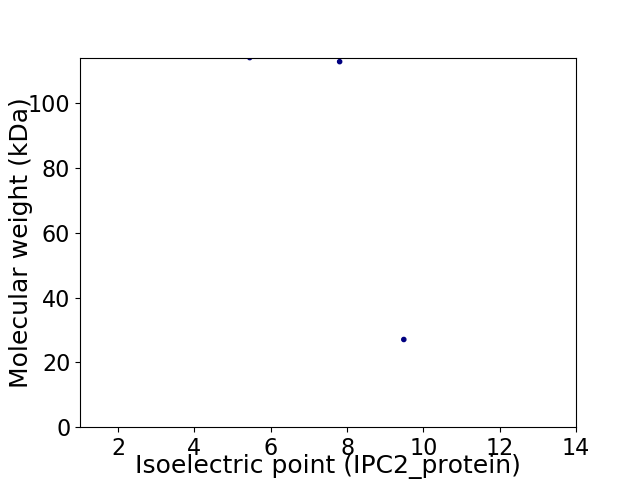
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
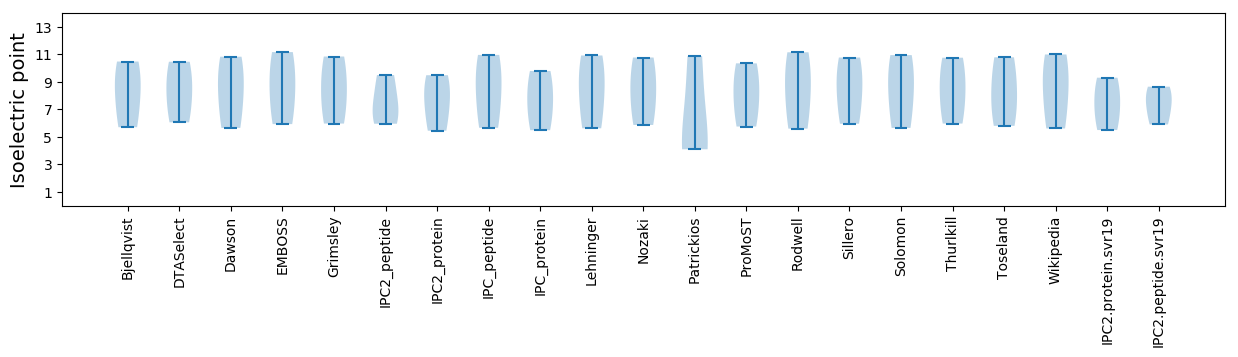
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q96724|POLS_DXV96 Structural polyprotein OS=Drosophila x virus (isolate Chung/1996) OX=654931 PE=1 SV=1
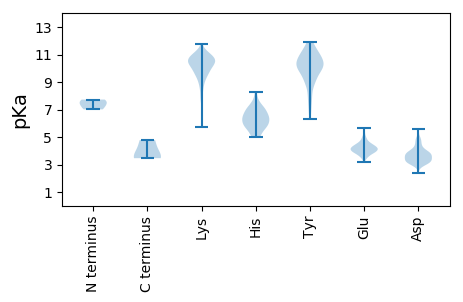
MM1 pKa = 7.01NTTNEE6 pKa = 4.03YY7 pKa = 10.89LKK9 pKa = 10.04TLLNPAQFISDD20 pKa = 3.74IPDD23 pKa = 4.8DD24 pKa = 3.62IMIRR28 pKa = 11.84HH29 pKa = 5.8VNSAQTITYY38 pKa = 8.65NLKK41 pKa = 10.44SGASGTGLIVVYY53 pKa = 9.91PNTPSSISGFHH64 pKa = 6.24YY65 pKa = 10.09IWDD68 pKa = 3.92SATSNWVFDD77 pKa = 3.51QYY79 pKa = 11.35IYY81 pKa = 10.05TAQEE85 pKa = 3.96LKK87 pKa = 10.76DD88 pKa = 3.6SYY90 pKa = 11.69DD91 pKa = 3.46YY92 pKa = 11.68GRR94 pKa = 11.84LISGSLSIKK103 pKa = 10.52SSTLPAGVYY112 pKa = 10.3ALNGTFNAVWFQGTLSEE129 pKa = 4.34VSDD132 pKa = 3.82YY133 pKa = 11.36SYY135 pKa = 11.85DD136 pKa = 4.06RR137 pKa = 11.84ILSITSNPLDD147 pKa = 3.22KK148 pKa = 11.14VGNVLVGDD156 pKa = 4.71GIEE159 pKa = 4.23VLSLPQGFNNPYY171 pKa = 9.84VRR173 pKa = 11.84LGDD176 pKa = 3.89KK177 pKa = 10.86SPSTLSSPTHH187 pKa = 5.61ITNTSQNLATGGAYY201 pKa = 8.02MIPVTTVPGQGFHH214 pKa = 6.53NKK216 pKa = 8.91EE217 pKa = 3.41FSINVDD223 pKa = 3.02SVGPVDD229 pKa = 4.5ILWSGQMTMQDD240 pKa = 3.61EE241 pKa = 4.59WTVTANYY248 pKa = 9.97QPLNISGTLIANSQRR263 pKa = 11.84TLTWSNTGVSNGSHH277 pKa = 5.22YY278 pKa = 11.12MNMNNLNVSLFHH290 pKa = 6.83EE291 pKa = 4.91NPPPEE296 pKa = 4.28PVAAIKK302 pKa = 10.66ININYY307 pKa = 9.06GNNTNGDD314 pKa = 3.49SSFSVDD320 pKa = 3.06SSFTINVIGGATIGVNSPTVGVGYY344 pKa = 10.46QGVAEE349 pKa = 4.54GTAITISGINNYY361 pKa = 9.17EE362 pKa = 4.12LVPNPDD368 pKa = 3.36LQKK371 pKa = 10.7NLPMTYY377 pKa = 8.93GTCDD381 pKa = 3.48PHH383 pKa = 8.25DD384 pKa = 3.86LTYY387 pKa = 10.63IKK389 pKa = 10.65YY390 pKa = 9.22ILSNRR395 pKa = 11.84EE396 pKa = 3.65QLGLRR401 pKa = 11.84SVMTLADD408 pKa = 3.91YY409 pKa = 11.54NRR411 pKa = 11.84MKK413 pKa = 10.18MYY415 pKa = 9.53MHH417 pKa = 7.05VLTNYY422 pKa = 10.13HH423 pKa = 4.97VDD425 pKa = 3.28EE426 pKa = 5.43RR427 pKa = 11.84EE428 pKa = 3.89ASSFDD433 pKa = 2.97FWQLLKK439 pKa = 10.56QIKK442 pKa = 9.58NVAVPLAATLAPQFAPIIGAADD464 pKa = 3.71GLANAILGDD473 pKa = 4.09SASGRR478 pKa = 11.84PVGNSASGMPISMSRR493 pKa = 11.84RR494 pKa = 11.84LRR496 pKa = 11.84NAYY499 pKa = 10.1SADD502 pKa = 3.53SPLGEE507 pKa = 4.32EE508 pKa = 3.44HH509 pKa = 7.09WLPNEE514 pKa = 3.89NEE516 pKa = 4.18NFNKK520 pKa = 9.81FDD522 pKa = 3.87IIYY525 pKa = 9.42DD526 pKa = 3.66VSHH529 pKa = 7.6SSMALFPVIMMEE541 pKa = 3.88HH542 pKa = 7.1DD543 pKa = 3.58KK544 pKa = 11.15VIPSDD549 pKa = 3.77PEE551 pKa = 3.51EE552 pKa = 4.86LYY554 pKa = 10.54IAVSLTEE561 pKa = 4.17SLRR564 pKa = 11.84KK565 pKa = 9.69QIPNLNDD572 pKa = 2.91MPYY575 pKa = 10.41YY576 pKa = 10.56EE577 pKa = 4.57MGGHH581 pKa = 5.96RR582 pKa = 11.84VYY584 pKa = 11.33NSVSSNVRR592 pKa = 11.84SGNFLRR598 pKa = 11.84SDD600 pKa = 4.3YY601 pKa = 11.03ILLPCYY607 pKa = 9.93QLLEE611 pKa = 4.28GRR613 pKa = 11.84LASSTSPNKK622 pKa = 9.21VTGTSHH628 pKa = 6.38QLAIYY633 pKa = 9.89AADD636 pKa = 4.45DD637 pKa = 3.97LLKK640 pKa = 11.07SGVLGKK646 pKa = 10.71APFAAFTGSVVGSSVGEE663 pKa = 3.75VFGINLKK670 pKa = 10.4LQLTDD675 pKa = 3.33SLGIPLLGNSPGLVQVKK692 pKa = 8.33TLTSLDD698 pKa = 3.76KK699 pKa = 10.7KK700 pKa = 10.23IKK702 pKa = 10.83DD703 pKa = 3.22MGDD706 pKa = 3.08VKK708 pKa = 10.77RR709 pKa = 11.84RR710 pKa = 11.84TPKK713 pKa = 8.76QTLPHH718 pKa = 5.61WTAGSASMNPFMNTNPFLEE737 pKa = 4.97EE738 pKa = 4.1LDD740 pKa = 3.66QPIPSNAAKK749 pKa = 10.02PISEE753 pKa = 4.24EE754 pKa = 3.98TRR756 pKa = 11.84DD757 pKa = 3.74LFLSDD762 pKa = 3.45GQTIPSSQEE771 pKa = 3.54KK772 pKa = 9.63IATIHH777 pKa = 6.63EE778 pKa = 4.3YY779 pKa = 10.91LLEE782 pKa = 4.54HH783 pKa = 6.96KK784 pKa = 9.81EE785 pKa = 3.86LEE787 pKa = 4.1EE788 pKa = 4.88AMFSLISQGRR798 pKa = 11.84GRR800 pKa = 11.84SLINMVVKK808 pKa = 10.54SALNIEE814 pKa = 4.43TQSRR818 pKa = 11.84EE819 pKa = 3.85VTGEE823 pKa = 3.19RR824 pKa = 11.84RR825 pKa = 11.84QRR827 pKa = 11.84LEE829 pKa = 3.63RR830 pKa = 11.84KK831 pKa = 9.19LRR833 pKa = 11.84NLEE836 pKa = 3.83NQGIYY841 pKa = 10.22VDD843 pKa = 3.85EE844 pKa = 4.46SKK846 pKa = 10.9IMSRR850 pKa = 11.84GRR852 pKa = 11.84ISKK855 pKa = 10.31EE856 pKa = 3.55DD857 pKa = 3.35TEE859 pKa = 4.37LAMRR863 pKa = 11.84IARR866 pKa = 11.84KK867 pKa = 7.61NQKK870 pKa = 9.05DD871 pKa = 3.23AKK873 pKa = 10.2LRR875 pKa = 11.84RR876 pKa = 11.84IYY878 pKa = 11.04SNNASIQEE886 pKa = 4.45SYY888 pKa = 10.35TVDD891 pKa = 3.8DD892 pKa = 4.99FVSYY896 pKa = 10.15WMEE899 pKa = 4.1QEE901 pKa = 4.19SLPTGIQIAMWLKK914 pKa = 10.72GDD916 pKa = 4.66DD917 pKa = 3.72WSQPIPPRR925 pKa = 11.84VQRR928 pKa = 11.84RR929 pKa = 11.84HH930 pKa = 5.12YY931 pKa = 10.67DD932 pKa = 3.13SYY934 pKa = 11.93IMMLGPSPTQEE945 pKa = 3.57QADD948 pKa = 3.94AVKK951 pKa = 10.9DD952 pKa = 3.99LVDD955 pKa = 5.6DD956 pKa = 4.59IYY958 pKa = 11.71DD959 pKa = 3.79RR960 pKa = 11.84NQGKK964 pKa = 9.79GPSQEE969 pKa = 3.94QARR972 pKa = 11.84EE973 pKa = 3.6LSHH976 pKa = 6.56AVRR979 pKa = 11.84RR980 pKa = 11.84LISHH984 pKa = 6.36SLVNQPATAPRR995 pKa = 11.84VPPRR999 pKa = 11.84RR1000 pKa = 11.84IVSAQTAQTDD1010 pKa = 3.89PPGRR1014 pKa = 11.84RR1015 pKa = 11.84AALDD1019 pKa = 3.04RR1020 pKa = 11.84LRR1022 pKa = 11.84RR1023 pKa = 11.84VRR1025 pKa = 11.84GEE1027 pKa = 4.23DD1028 pKa = 3.12NDD1030 pKa = 3.87IVV1032 pKa = 3.53
MM1 pKa = 7.01NTTNEE6 pKa = 4.03YY7 pKa = 10.89LKK9 pKa = 10.04TLLNPAQFISDD20 pKa = 3.74IPDD23 pKa = 4.8DD24 pKa = 3.62IMIRR28 pKa = 11.84HH29 pKa = 5.8VNSAQTITYY38 pKa = 8.65NLKK41 pKa = 10.44SGASGTGLIVVYY53 pKa = 9.91PNTPSSISGFHH64 pKa = 6.24YY65 pKa = 10.09IWDD68 pKa = 3.92SATSNWVFDD77 pKa = 3.51QYY79 pKa = 11.35IYY81 pKa = 10.05TAQEE85 pKa = 3.96LKK87 pKa = 10.76DD88 pKa = 3.6SYY90 pKa = 11.69DD91 pKa = 3.46YY92 pKa = 11.68GRR94 pKa = 11.84LISGSLSIKK103 pKa = 10.52SSTLPAGVYY112 pKa = 10.3ALNGTFNAVWFQGTLSEE129 pKa = 4.34VSDD132 pKa = 3.82YY133 pKa = 11.36SYY135 pKa = 11.85DD136 pKa = 4.06RR137 pKa = 11.84ILSITSNPLDD147 pKa = 3.22KK148 pKa = 11.14VGNVLVGDD156 pKa = 4.71GIEE159 pKa = 4.23VLSLPQGFNNPYY171 pKa = 9.84VRR173 pKa = 11.84LGDD176 pKa = 3.89KK177 pKa = 10.86SPSTLSSPTHH187 pKa = 5.61ITNTSQNLATGGAYY201 pKa = 8.02MIPVTTVPGQGFHH214 pKa = 6.53NKK216 pKa = 8.91EE217 pKa = 3.41FSINVDD223 pKa = 3.02SVGPVDD229 pKa = 4.5ILWSGQMTMQDD240 pKa = 3.61EE241 pKa = 4.59WTVTANYY248 pKa = 9.97QPLNISGTLIANSQRR263 pKa = 11.84TLTWSNTGVSNGSHH277 pKa = 5.22YY278 pKa = 11.12MNMNNLNVSLFHH290 pKa = 6.83EE291 pKa = 4.91NPPPEE296 pKa = 4.28PVAAIKK302 pKa = 10.66ININYY307 pKa = 9.06GNNTNGDD314 pKa = 3.49SSFSVDD320 pKa = 3.06SSFTINVIGGATIGVNSPTVGVGYY344 pKa = 10.46QGVAEE349 pKa = 4.54GTAITISGINNYY361 pKa = 9.17EE362 pKa = 4.12LVPNPDD368 pKa = 3.36LQKK371 pKa = 10.7NLPMTYY377 pKa = 8.93GTCDD381 pKa = 3.48PHH383 pKa = 8.25DD384 pKa = 3.86LTYY387 pKa = 10.63IKK389 pKa = 10.65YY390 pKa = 9.22ILSNRR395 pKa = 11.84EE396 pKa = 3.65QLGLRR401 pKa = 11.84SVMTLADD408 pKa = 3.91YY409 pKa = 11.54NRR411 pKa = 11.84MKK413 pKa = 10.18MYY415 pKa = 9.53MHH417 pKa = 7.05VLTNYY422 pKa = 10.13HH423 pKa = 4.97VDD425 pKa = 3.28EE426 pKa = 5.43RR427 pKa = 11.84EE428 pKa = 3.89ASSFDD433 pKa = 2.97FWQLLKK439 pKa = 10.56QIKK442 pKa = 9.58NVAVPLAATLAPQFAPIIGAADD464 pKa = 3.71GLANAILGDD473 pKa = 4.09SASGRR478 pKa = 11.84PVGNSASGMPISMSRR493 pKa = 11.84RR494 pKa = 11.84LRR496 pKa = 11.84NAYY499 pKa = 10.1SADD502 pKa = 3.53SPLGEE507 pKa = 4.32EE508 pKa = 3.44HH509 pKa = 7.09WLPNEE514 pKa = 3.89NEE516 pKa = 4.18NFNKK520 pKa = 9.81FDD522 pKa = 3.87IIYY525 pKa = 9.42DD526 pKa = 3.66VSHH529 pKa = 7.6SSMALFPVIMMEE541 pKa = 3.88HH542 pKa = 7.1DD543 pKa = 3.58KK544 pKa = 11.15VIPSDD549 pKa = 3.77PEE551 pKa = 3.51EE552 pKa = 4.86LYY554 pKa = 10.54IAVSLTEE561 pKa = 4.17SLRR564 pKa = 11.84KK565 pKa = 9.69QIPNLNDD572 pKa = 2.91MPYY575 pKa = 10.41YY576 pKa = 10.56EE577 pKa = 4.57MGGHH581 pKa = 5.96RR582 pKa = 11.84VYY584 pKa = 11.33NSVSSNVRR592 pKa = 11.84SGNFLRR598 pKa = 11.84SDD600 pKa = 4.3YY601 pKa = 11.03ILLPCYY607 pKa = 9.93QLLEE611 pKa = 4.28GRR613 pKa = 11.84LASSTSPNKK622 pKa = 9.21VTGTSHH628 pKa = 6.38QLAIYY633 pKa = 9.89AADD636 pKa = 4.45DD637 pKa = 3.97LLKK640 pKa = 11.07SGVLGKK646 pKa = 10.71APFAAFTGSVVGSSVGEE663 pKa = 3.75VFGINLKK670 pKa = 10.4LQLTDD675 pKa = 3.33SLGIPLLGNSPGLVQVKK692 pKa = 8.33TLTSLDD698 pKa = 3.76KK699 pKa = 10.7KK700 pKa = 10.23IKK702 pKa = 10.83DD703 pKa = 3.22MGDD706 pKa = 3.08VKK708 pKa = 10.77RR709 pKa = 11.84RR710 pKa = 11.84TPKK713 pKa = 8.76QTLPHH718 pKa = 5.61WTAGSASMNPFMNTNPFLEE737 pKa = 4.97EE738 pKa = 4.1LDD740 pKa = 3.66QPIPSNAAKK749 pKa = 10.02PISEE753 pKa = 4.24EE754 pKa = 3.98TRR756 pKa = 11.84DD757 pKa = 3.74LFLSDD762 pKa = 3.45GQTIPSSQEE771 pKa = 3.54KK772 pKa = 9.63IATIHH777 pKa = 6.63EE778 pKa = 4.3YY779 pKa = 10.91LLEE782 pKa = 4.54HH783 pKa = 6.96KK784 pKa = 9.81EE785 pKa = 3.86LEE787 pKa = 4.1EE788 pKa = 4.88AMFSLISQGRR798 pKa = 11.84GRR800 pKa = 11.84SLINMVVKK808 pKa = 10.54SALNIEE814 pKa = 4.43TQSRR818 pKa = 11.84EE819 pKa = 3.85VTGEE823 pKa = 3.19RR824 pKa = 11.84RR825 pKa = 11.84QRR827 pKa = 11.84LEE829 pKa = 3.63RR830 pKa = 11.84KK831 pKa = 9.19LRR833 pKa = 11.84NLEE836 pKa = 3.83NQGIYY841 pKa = 10.22VDD843 pKa = 3.85EE844 pKa = 4.46SKK846 pKa = 10.9IMSRR850 pKa = 11.84GRR852 pKa = 11.84ISKK855 pKa = 10.31EE856 pKa = 3.55DD857 pKa = 3.35TEE859 pKa = 4.37LAMRR863 pKa = 11.84IARR866 pKa = 11.84KK867 pKa = 7.61NQKK870 pKa = 9.05DD871 pKa = 3.23AKK873 pKa = 10.2LRR875 pKa = 11.84RR876 pKa = 11.84IYY878 pKa = 11.04SNNASIQEE886 pKa = 4.45SYY888 pKa = 10.35TVDD891 pKa = 3.8DD892 pKa = 4.99FVSYY896 pKa = 10.15WMEE899 pKa = 4.1QEE901 pKa = 4.19SLPTGIQIAMWLKK914 pKa = 10.72GDD916 pKa = 4.66DD917 pKa = 3.72WSQPIPPRR925 pKa = 11.84VQRR928 pKa = 11.84RR929 pKa = 11.84HH930 pKa = 5.12YY931 pKa = 10.67DD932 pKa = 3.13SYY934 pKa = 11.93IMMLGPSPTQEE945 pKa = 3.57QADD948 pKa = 3.94AVKK951 pKa = 10.9DD952 pKa = 3.99LVDD955 pKa = 5.6DD956 pKa = 4.59IYY958 pKa = 11.71DD959 pKa = 3.79RR960 pKa = 11.84NQGKK964 pKa = 9.79GPSQEE969 pKa = 3.94QARR972 pKa = 11.84EE973 pKa = 3.6LSHH976 pKa = 6.56AVRR979 pKa = 11.84RR980 pKa = 11.84LISHH984 pKa = 6.36SLVNQPATAPRR995 pKa = 11.84VPPRR999 pKa = 11.84RR1000 pKa = 11.84IVSAQTAQTDD1010 pKa = 3.89PPGRR1014 pKa = 11.84RR1015 pKa = 11.84AALDD1019 pKa = 3.04RR1020 pKa = 11.84LRR1022 pKa = 11.84RR1023 pKa = 11.84VRR1025 pKa = 11.84GEE1027 pKa = 4.23DD1028 pKa = 3.12NDD1030 pKa = 3.87IVV1032 pKa = 3.53
Molecular weight: 114.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q91CD5|RDRP_DXV96 RNA-directed RNA polymerase OS=Drosophila x virus (isolate Chung/1996) OX=654931 GN=VP1 PE=3 SV=2
MM1 pKa = 7.44LSIIRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 7.68TRR10 pKa = 11.84IVDD13 pKa = 3.05ITKK16 pKa = 10.21QGNGNVPPACHH27 pKa = 6.01LCSRR31 pKa = 11.84RR32 pKa = 11.84LTKK35 pKa = 9.88EE36 pKa = 3.27WSFGEE41 pKa = 4.05GTICSFHH48 pKa = 7.63RR49 pKa = 11.84IRR51 pKa = 11.84CRR53 pKa = 11.84VQCGRR58 pKa = 11.84SFWHH62 pKa = 6.17QPEE65 pKa = 4.26TPIDD69 pKa = 3.95GLSWDD74 pKa = 3.71STTWEE79 pKa = 4.11LTRR82 pKa = 11.84ISSSKK87 pKa = 11.04DD88 pKa = 3.02PDD90 pKa = 3.12ISGQKK95 pKa = 9.53DD96 pKa = 2.87KK97 pKa = 11.13RR98 pKa = 11.84YY99 pKa = 10.13GRR101 pKa = 11.84RR102 pKa = 11.84KK103 pKa = 9.38EE104 pKa = 4.22KK105 pKa = 10.68NPKK108 pKa = 8.49TDD110 pKa = 3.52PPALDD115 pKa = 3.27SRR117 pKa = 11.84VRR119 pKa = 11.84EE120 pKa = 3.98HH121 pKa = 6.9EE122 pKa = 4.75PIHH125 pKa = 5.07EE126 pKa = 4.48HH127 pKa = 5.86EE128 pKa = 5.36PIPGRR133 pKa = 11.84VGPADD138 pKa = 3.28TKK140 pKa = 10.48QRR142 pKa = 11.84CKK144 pKa = 11.15ANLRR148 pKa = 11.84GDD150 pKa = 3.65SGFVSIGRR158 pKa = 11.84SNHH161 pKa = 5.93PKK163 pKa = 10.47LSRR166 pKa = 11.84EE167 pKa = 4.08DD168 pKa = 3.56CHH170 pKa = 5.68NTRR173 pKa = 11.84VPPGTQGVRR182 pKa = 11.84GGNVQLDD189 pKa = 4.03KK190 pKa = 10.98PRR192 pKa = 11.84EE193 pKa = 3.97RR194 pKa = 11.84PVSYY198 pKa = 10.21QHH200 pKa = 6.48GSKK203 pKa = 10.11KK204 pKa = 10.17RR205 pKa = 11.84SEE207 pKa = 4.11HH208 pKa = 6.25RR209 pKa = 11.84NPVSRR214 pKa = 11.84SHH216 pKa = 6.41RR217 pKa = 11.84RR218 pKa = 11.84KK219 pKa = 9.46KK220 pKa = 10.73AKK222 pKa = 9.43TRR224 pKa = 11.84TKK226 pKa = 9.67TSKK229 pKa = 10.24LGKK232 pKa = 10.21SRR234 pKa = 11.84DD235 pKa = 3.24ICC237 pKa = 4.78
MM1 pKa = 7.44LSIIRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 7.68TRR10 pKa = 11.84IVDD13 pKa = 3.05ITKK16 pKa = 10.21QGNGNVPPACHH27 pKa = 6.01LCSRR31 pKa = 11.84RR32 pKa = 11.84LTKK35 pKa = 9.88EE36 pKa = 3.27WSFGEE41 pKa = 4.05GTICSFHH48 pKa = 7.63RR49 pKa = 11.84IRR51 pKa = 11.84CRR53 pKa = 11.84VQCGRR58 pKa = 11.84SFWHH62 pKa = 6.17QPEE65 pKa = 4.26TPIDD69 pKa = 3.95GLSWDD74 pKa = 3.71STTWEE79 pKa = 4.11LTRR82 pKa = 11.84ISSSKK87 pKa = 11.04DD88 pKa = 3.02PDD90 pKa = 3.12ISGQKK95 pKa = 9.53DD96 pKa = 2.87KK97 pKa = 11.13RR98 pKa = 11.84YY99 pKa = 10.13GRR101 pKa = 11.84RR102 pKa = 11.84KK103 pKa = 9.38EE104 pKa = 4.22KK105 pKa = 10.68NPKK108 pKa = 8.49TDD110 pKa = 3.52PPALDD115 pKa = 3.27SRR117 pKa = 11.84VRR119 pKa = 11.84EE120 pKa = 3.98HH121 pKa = 6.9EE122 pKa = 4.75PIHH125 pKa = 5.07EE126 pKa = 4.48HH127 pKa = 5.86EE128 pKa = 5.36PIPGRR133 pKa = 11.84VGPADD138 pKa = 3.28TKK140 pKa = 10.48QRR142 pKa = 11.84CKK144 pKa = 11.15ANLRR148 pKa = 11.84GDD150 pKa = 3.65SGFVSIGRR158 pKa = 11.84SNHH161 pKa = 5.93PKK163 pKa = 10.47LSRR166 pKa = 11.84EE167 pKa = 4.08DD168 pKa = 3.56CHH170 pKa = 5.68NTRR173 pKa = 11.84VPPGTQGVRR182 pKa = 11.84GGNVQLDD189 pKa = 4.03KK190 pKa = 10.98PRR192 pKa = 11.84EE193 pKa = 3.97RR194 pKa = 11.84PVSYY198 pKa = 10.21QHH200 pKa = 6.48GSKK203 pKa = 10.11KK204 pKa = 10.17RR205 pKa = 11.84SEE207 pKa = 4.11HH208 pKa = 6.25RR209 pKa = 11.84NPVSRR214 pKa = 11.84SHH216 pKa = 6.41RR217 pKa = 11.84RR218 pKa = 11.84KK219 pKa = 9.46KK220 pKa = 10.73AKK222 pKa = 9.43TRR224 pKa = 11.84TKK226 pKa = 9.67TSKK229 pKa = 10.24LGKK232 pKa = 10.21SRR234 pKa = 11.84DD235 pKa = 3.24ICC237 pKa = 4.78
Molecular weight: 27.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2266 |
237 |
1032 |
755.3 |
84.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.781 ± 0.786 | 0.574 ± 0.594 |
5.384 ± 0.149 | 5.56 ± 0.531 |
2.295 ± 0.153 | 6.311 ± 0.597 |
1.986 ± 0.594 | 6.134 ± 0.305 |
6.002 ± 1.237 | 8.738 ± 0.994 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.692 ± 0.485 | 5.869 ± 0.659 |
6.134 ± 0.31 | 4.016 ± 0.203 |
5.605 ± 1.601 | 8.914 ± 0.769 |
6.399 ± 0.18 | 6.046 ± 0.3 |
1.412 ± 0.138 | 4.148 ± 0.794 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
