
Cuban alphasatellite 1
Taxonomy: Viruses; Alphasatellitidae; Geminialphasatellitinae; unclassified Begomovirus-associated alphasatellites
Average proteome isoelectric point is 7.68
Get precalculated fractions of proteins
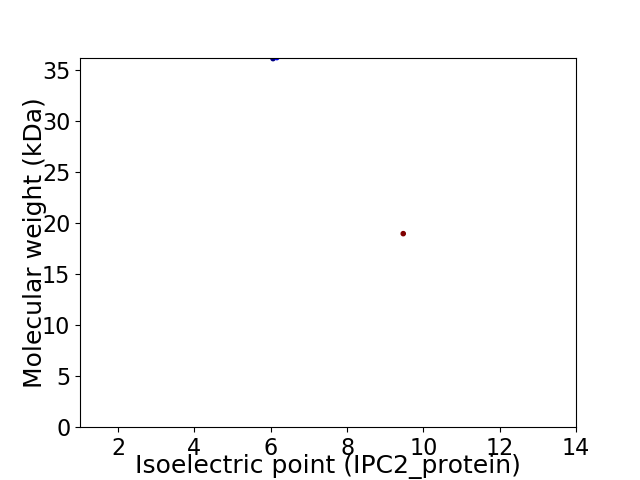
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|S6CRB1|S6CRB1_9VIRU Rep OS=Cuban alphasatellite 1 OX=1188819 GN=V1 PE=4 SV=1
MM1 pKa = 7.51AKK3 pKa = 9.94SRR5 pKa = 11.84NYY7 pKa = 10.4CFTLFSYY14 pKa = 9.58VLPLFNSLPDD24 pKa = 3.31WANYY28 pKa = 10.28LVFQEE33 pKa = 4.78EE34 pKa = 4.64EE35 pKa = 4.28SPSTGRR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.55IQGYY47 pKa = 10.2VNLKK51 pKa = 9.96SPQSFSFLKK60 pKa = 10.5KK61 pKa = 9.76KK62 pKa = 10.78LGDD65 pKa = 3.86GVHH68 pKa = 6.76LEE70 pKa = 4.03QARR73 pKa = 11.84GSASCNRR80 pKa = 11.84DD81 pKa = 3.05YY82 pKa = 11.22CRR84 pKa = 11.84KK85 pKa = 7.55TDD87 pKa = 3.52SRR89 pKa = 11.84VSGPWEE95 pKa = 3.82FGILAEE101 pKa = 4.33QGSKK105 pKa = 9.95KK106 pKa = 10.06RR107 pKa = 11.84KK108 pKa = 7.07TMEE111 pKa = 4.01SFQEE115 pKa = 4.17DD116 pKa = 4.27PEE118 pKa = 4.18EE119 pKa = 4.99LRR121 pKa = 11.84LSDD124 pKa = 3.31PKK126 pKa = 10.91LYY128 pKa = 10.27RR129 pKa = 11.84RR130 pKa = 11.84CLATRR135 pKa = 11.84VNTEE139 pKa = 3.42FAGLVLPVLDD149 pKa = 5.35RR150 pKa = 11.84PWQLLVEE157 pKa = 4.47KK158 pKa = 10.54VLDD161 pKa = 4.18EE162 pKa = 4.9GPDD165 pKa = 3.38DD166 pKa = 3.88RR167 pKa = 11.84TIIWVYY173 pKa = 10.56GSQGNEE179 pKa = 4.34GKK181 pKa = 7.1TTWAKK186 pKa = 10.81SKK188 pKa = 10.64VQAGWFYY195 pKa = 11.16SRR197 pKa = 11.84GGKK200 pKa = 9.86GEE202 pKa = 4.16NIKK205 pKa = 10.8YY206 pKa = 10.46SYY208 pKa = 11.0AEE210 pKa = 4.1HH211 pKa = 6.89LGHH214 pKa = 7.21AVFDD218 pKa = 3.76IPRR221 pKa = 11.84QVEE224 pKa = 4.18DD225 pKa = 3.31VLQYY229 pKa = 10.56TVLEE233 pKa = 4.5EE234 pKa = 3.95IKK236 pKa = 10.78DD237 pKa = 3.69RR238 pKa = 11.84LIRR241 pKa = 11.84SSKK244 pKa = 10.56YY245 pKa = 10.02EE246 pKa = 4.51PIDD249 pKa = 3.92FNCSDD254 pKa = 3.5QVHH257 pKa = 5.25VVVLSNFLPQLDD269 pKa = 4.43SEE271 pKa = 4.5HH272 pKa = 7.42DD273 pKa = 3.61SRR275 pKa = 11.84GNLIKK280 pKa = 10.61KK281 pKa = 9.93QMLSRR286 pKa = 11.84DD287 pKa = 2.9RR288 pKa = 11.84VVIVNIAEE296 pKa = 4.15SLIVRR301 pKa = 11.84DD302 pKa = 3.66NEE304 pKa = 4.23TVSFHH309 pKa = 7.24EE310 pKa = 4.25YY311 pKa = 9.9MEE313 pKa = 4.37
MM1 pKa = 7.51AKK3 pKa = 9.94SRR5 pKa = 11.84NYY7 pKa = 10.4CFTLFSYY14 pKa = 9.58VLPLFNSLPDD24 pKa = 3.31WANYY28 pKa = 10.28LVFQEE33 pKa = 4.78EE34 pKa = 4.64EE35 pKa = 4.28SPSTGRR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.55IQGYY47 pKa = 10.2VNLKK51 pKa = 9.96SPQSFSFLKK60 pKa = 10.5KK61 pKa = 9.76KK62 pKa = 10.78LGDD65 pKa = 3.86GVHH68 pKa = 6.76LEE70 pKa = 4.03QARR73 pKa = 11.84GSASCNRR80 pKa = 11.84DD81 pKa = 3.05YY82 pKa = 11.22CRR84 pKa = 11.84KK85 pKa = 7.55TDD87 pKa = 3.52SRR89 pKa = 11.84VSGPWEE95 pKa = 3.82FGILAEE101 pKa = 4.33QGSKK105 pKa = 9.95KK106 pKa = 10.06RR107 pKa = 11.84KK108 pKa = 7.07TMEE111 pKa = 4.01SFQEE115 pKa = 4.17DD116 pKa = 4.27PEE118 pKa = 4.18EE119 pKa = 4.99LRR121 pKa = 11.84LSDD124 pKa = 3.31PKK126 pKa = 10.91LYY128 pKa = 10.27RR129 pKa = 11.84RR130 pKa = 11.84CLATRR135 pKa = 11.84VNTEE139 pKa = 3.42FAGLVLPVLDD149 pKa = 5.35RR150 pKa = 11.84PWQLLVEE157 pKa = 4.47KK158 pKa = 10.54VLDD161 pKa = 4.18EE162 pKa = 4.9GPDD165 pKa = 3.38DD166 pKa = 3.88RR167 pKa = 11.84TIIWVYY173 pKa = 10.56GSQGNEE179 pKa = 4.34GKK181 pKa = 7.1TTWAKK186 pKa = 10.81SKK188 pKa = 10.64VQAGWFYY195 pKa = 11.16SRR197 pKa = 11.84GGKK200 pKa = 9.86GEE202 pKa = 4.16NIKK205 pKa = 10.8YY206 pKa = 10.46SYY208 pKa = 11.0AEE210 pKa = 4.1HH211 pKa = 6.89LGHH214 pKa = 7.21AVFDD218 pKa = 3.76IPRR221 pKa = 11.84QVEE224 pKa = 4.18DD225 pKa = 3.31VLQYY229 pKa = 10.56TVLEE233 pKa = 4.5EE234 pKa = 3.95IKK236 pKa = 10.78DD237 pKa = 3.69RR238 pKa = 11.84LIRR241 pKa = 11.84SSKK244 pKa = 10.56YY245 pKa = 10.02EE246 pKa = 4.51PIDD249 pKa = 3.92FNCSDD254 pKa = 3.5QVHH257 pKa = 5.25VVVLSNFLPQLDD269 pKa = 4.43SEE271 pKa = 4.5HH272 pKa = 7.42DD273 pKa = 3.61SRR275 pKa = 11.84GNLIKK280 pKa = 10.61KK281 pKa = 9.93QMLSRR286 pKa = 11.84DD287 pKa = 2.9RR288 pKa = 11.84VVIVNIAEE296 pKa = 4.15SLIVRR301 pKa = 11.84DD302 pKa = 3.66NEE304 pKa = 4.23TVSFHH309 pKa = 7.24EE310 pKa = 4.25YY311 pKa = 9.9MEE313 pKa = 4.37
Molecular weight: 36.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S6CRB1|S6CRB1_9VIRU Rep OS=Cuban alphasatellite 1 OX=1188819 GN=V1 PE=4 SV=1
MM1 pKa = 7.41GFISNKK7 pKa = 10.1PGDD10 pKa = 4.05LHH12 pKa = 6.36PAIEE16 pKa = 4.88IIAEE20 pKa = 4.11RR21 pKa = 11.84LTLASLGHH29 pKa = 6.41GSSAYY34 pKa = 10.16LRR36 pKa = 11.84NRR38 pKa = 11.84GARR41 pKa = 11.84RR42 pKa = 11.84GRR44 pKa = 11.84RR45 pKa = 11.84WNHH48 pKa = 4.29FRR50 pKa = 11.84KK51 pKa = 7.52TQRR54 pKa = 11.84NYY56 pKa = 11.26DD57 pKa = 3.96CPTLNSIVAAWRR69 pKa = 11.84LGLIRR74 pKa = 11.84SSLVWYY80 pKa = 10.23SLYY83 pKa = 10.2LTDD86 pKa = 6.02HH87 pKa = 6.43GSCWLRR93 pKa = 11.84RR94 pKa = 11.84YY95 pKa = 7.29STRR98 pKa = 11.84AQMIEE103 pKa = 3.86LSYY106 pKa = 11.27GCMALRR112 pKa = 11.84VMKK115 pKa = 10.58GKK117 pKa = 10.66LPGQSLRR124 pKa = 11.84FKK126 pKa = 10.97LGGSTLEE133 pKa = 4.06EE134 pKa = 4.22EE135 pKa = 4.48KK136 pKa = 10.88EE137 pKa = 4.39KK138 pKa = 10.57TSSTHH143 pKa = 5.1TLNTSAMLCSTSHH156 pKa = 6.5AKK158 pKa = 9.21WKK160 pKa = 8.08TFCNTLCC167 pKa = 4.58
MM1 pKa = 7.41GFISNKK7 pKa = 10.1PGDD10 pKa = 4.05LHH12 pKa = 6.36PAIEE16 pKa = 4.88IIAEE20 pKa = 4.11RR21 pKa = 11.84LTLASLGHH29 pKa = 6.41GSSAYY34 pKa = 10.16LRR36 pKa = 11.84NRR38 pKa = 11.84GARR41 pKa = 11.84RR42 pKa = 11.84GRR44 pKa = 11.84RR45 pKa = 11.84WNHH48 pKa = 4.29FRR50 pKa = 11.84KK51 pKa = 7.52TQRR54 pKa = 11.84NYY56 pKa = 11.26DD57 pKa = 3.96CPTLNSIVAAWRR69 pKa = 11.84LGLIRR74 pKa = 11.84SSLVWYY80 pKa = 10.23SLYY83 pKa = 10.2LTDD86 pKa = 6.02HH87 pKa = 6.43GSCWLRR93 pKa = 11.84RR94 pKa = 11.84YY95 pKa = 7.29STRR98 pKa = 11.84AQMIEE103 pKa = 3.86LSYY106 pKa = 11.27GCMALRR112 pKa = 11.84VMKK115 pKa = 10.58GKK117 pKa = 10.66LPGQSLRR124 pKa = 11.84FKK126 pKa = 10.97LGGSTLEE133 pKa = 4.06EE134 pKa = 4.22EE135 pKa = 4.48KK136 pKa = 10.88EE137 pKa = 4.39KK138 pKa = 10.57TSSTHH143 pKa = 5.1TLNTSAMLCSTSHH156 pKa = 6.5AKK158 pKa = 9.21WKK160 pKa = 8.08TFCNTLCC167 pKa = 4.58
Molecular weight: 18.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
480 |
167 |
313 |
240.0 |
27.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.792 ± 1.006 | 2.292 ± 0.729 |
4.583 ± 1.562 | 6.667 ± 1.387 |
3.75 ± 0.759 | 6.667 ± 0.626 |
2.708 ± 0.496 | 4.167 ± 0.014 |
6.042 ± 0.366 | 10.625 ± 1.093 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.875 ± 0.627 | 4.167 ± 0.014 |
3.542 ± 0.643 | 3.542 ± 0.978 |
7.917 ± 0.933 | 9.583 ± 0.67 |
5.0 ± 1.56 | 5.833 ± 2.262 |
2.292 ± 0.394 | 3.958 ± 0.205 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
