
Circovirus-like genome CB-B
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.53
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
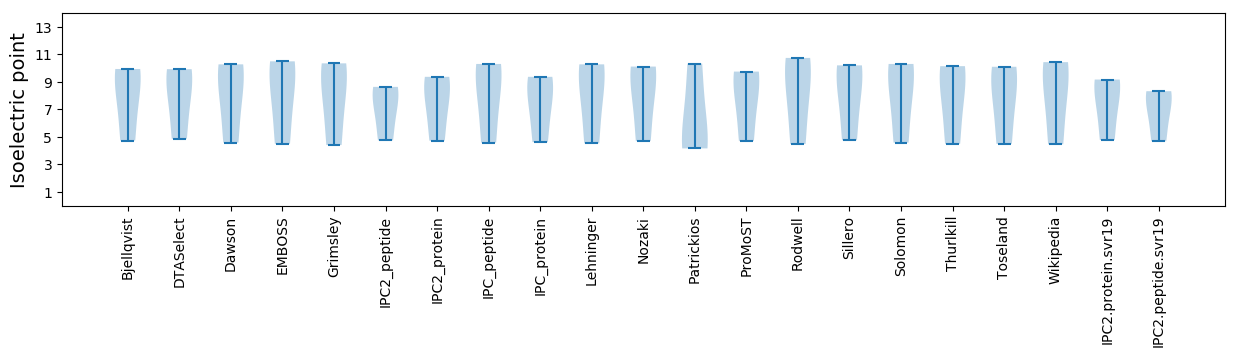
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6GII9|C6GII9_9VIRU Putative Rep-associated protein OS=Circovirus-like genome CB-B OX=642257 PE=4 SV=1
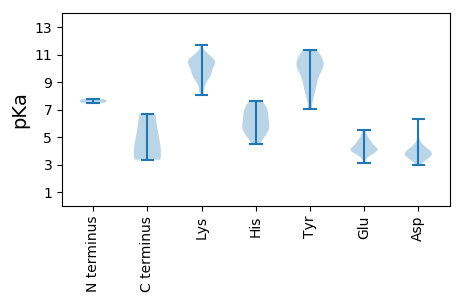
MM1 pKa = 7.62LATPPNSPTVRR12 pKa = 11.84HH13 pKa = 5.65RR14 pKa = 11.84HH15 pKa = 5.12QGPAGDD21 pKa = 4.23SPDD24 pKa = 3.79NGVQALASGRR34 pKa = 11.84QAQEE38 pKa = 3.09QAAAFWVEE46 pKa = 3.94SSFPAGDD53 pKa = 3.35PRR55 pKa = 11.84RR56 pKa = 11.84AYY58 pKa = 9.93EE59 pKa = 3.9EE60 pKa = 3.82QRR62 pKa = 11.84SVGLCGEE69 pKa = 4.53GGDD72 pKa = 3.9FRR74 pKa = 11.84RR75 pKa = 11.84WDD77 pKa = 3.73EE78 pKa = 4.08LPAGEE83 pKa = 4.78GAKK86 pKa = 9.97QGSGLGPSEE95 pKa = 4.7GPSSEE100 pKa = 4.16GPVLGDD106 pKa = 4.14SLWHH110 pKa = 5.97AAQVRR115 pKa = 11.84RR116 pKa = 11.84LWEE119 pKa = 3.93MTDD122 pKa = 3.7VMWGDD127 pKa = 4.14LGMDD131 pKa = 4.09TDD133 pKa = 4.39PAHH136 pKa = 6.69
MM1 pKa = 7.62LATPPNSPTVRR12 pKa = 11.84HH13 pKa = 5.65RR14 pKa = 11.84HH15 pKa = 5.12QGPAGDD21 pKa = 4.23SPDD24 pKa = 3.79NGVQALASGRR34 pKa = 11.84QAQEE38 pKa = 3.09QAAAFWVEE46 pKa = 3.94SSFPAGDD53 pKa = 3.35PRR55 pKa = 11.84RR56 pKa = 11.84AYY58 pKa = 9.93EE59 pKa = 3.9EE60 pKa = 3.82QRR62 pKa = 11.84SVGLCGEE69 pKa = 4.53GGDD72 pKa = 3.9FRR74 pKa = 11.84RR75 pKa = 11.84WDD77 pKa = 3.73EE78 pKa = 4.08LPAGEE83 pKa = 4.78GAKK86 pKa = 9.97QGSGLGPSEE95 pKa = 4.7GPSSEE100 pKa = 4.16GPVLGDD106 pKa = 4.14SLWHH110 pKa = 5.97AAQVRR115 pKa = 11.84RR116 pKa = 11.84LWEE119 pKa = 3.93MTDD122 pKa = 3.7VMWGDD127 pKa = 4.14LGMDD131 pKa = 4.09TDD133 pKa = 4.39PAHH136 pKa = 6.69
Molecular weight: 14.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6GII8|C6GII8_9VIRU Rep-associated protein OS=Circovirus-like genome CB-B OX=642257 PE=4 SV=1
MM1 pKa = 7.77CIFPVSLLSKK11 pKa = 10.7KK12 pKa = 10.4NINCTMVVWPAVLPYY27 pKa = 10.57LEE29 pKa = 4.79AANTVAQGVQTANTIVHH46 pKa = 6.95EE47 pKa = 4.62GQQLAKK53 pKa = 9.47RR54 pKa = 11.84TFGTQISNKK63 pKa = 8.24NKK65 pKa = 9.46KK66 pKa = 9.24RR67 pKa = 11.84KK68 pKa = 9.59LDD70 pKa = 3.46VGHH73 pKa = 7.61ASTQTSGGGKK83 pKa = 9.57YY84 pKa = 9.29YY85 pKa = 9.92QAGYY89 pKa = 9.8VGRR92 pKa = 11.84FRR94 pKa = 11.84GKK96 pKa = 8.34RR97 pKa = 11.84AKK99 pKa = 9.43IVRR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84IPSKK109 pKa = 10.51KK110 pKa = 9.32RR111 pKa = 11.84VAKK114 pKa = 10.23KK115 pKa = 9.59RR116 pKa = 11.84RR117 pKa = 11.84TYY119 pKa = 9.99KK120 pKa = 10.17RR121 pKa = 11.84KK122 pKa = 8.79SRR124 pKa = 11.84VVKK127 pKa = 10.53GEE129 pKa = 3.63NYY131 pKa = 8.67YY132 pKa = 11.27GRR134 pKa = 11.84VGSIATLEE142 pKa = 4.22VTGALSDD149 pKa = 3.78SDD151 pKa = 3.93CVYY154 pKa = 10.42IGHH157 pKa = 6.55SSFAVGQVMRR167 pKa = 11.84MLSVAIMRR175 pKa = 11.84KK176 pKa = 8.09LFKK179 pKa = 9.97MAISWEE185 pKa = 4.06PEE187 pKa = 4.01NISTNIPYY195 pKa = 10.44KK196 pKa = 10.59LVGGAQVSNGYY207 pKa = 7.05TLIVKK212 pKa = 9.99KK213 pKa = 10.71INTDD217 pKa = 3.89DD218 pKa = 3.66NVKK221 pKa = 8.73TQDD224 pKa = 3.64EE225 pKa = 5.16YY226 pKa = 10.9IGPANSHH233 pKa = 6.78NITAPADD240 pKa = 3.55WFRR243 pKa = 11.84TNHH246 pKa = 5.49MEE248 pKa = 5.19AISAQASSNTAVEE261 pKa = 4.3RR262 pKa = 11.84NNRR265 pKa = 11.84LLWLHH270 pKa = 6.17LVDD273 pKa = 3.73VGTGIVRR280 pKa = 11.84AQLDD284 pKa = 4.72LTTLMCDD291 pKa = 3.42VYY293 pKa = 11.33SKK295 pKa = 11.69SNLKK299 pKa = 9.04IQNVTVPSATATEE312 pKa = 4.04ADD314 pKa = 3.44NVNNVPLIGRR324 pKa = 11.84SYY326 pKa = 10.51HH327 pKa = 5.55ISNWQPMTADD337 pKa = 4.37DD338 pKa = 6.29DD339 pKa = 4.06MGPLNRR345 pKa = 11.84LNQDD349 pKa = 3.28TGVLGVEE356 pKa = 4.9IKK358 pKa = 10.65QSATLGSQYY367 pKa = 8.53VTWKK371 pKa = 10.25EE372 pKa = 4.0PPPSKK377 pKa = 10.97AFINCVSSSPEE388 pKa = 3.54RR389 pKa = 11.84MEE391 pKa = 4.65PGTIKK396 pKa = 10.61NHH398 pKa = 4.52VSILRR403 pKa = 11.84RR404 pKa = 11.84SMAFPRR410 pKa = 11.84LMEE413 pKa = 4.33ALAIRR418 pKa = 11.84RR419 pKa = 11.84GNVLNKK425 pKa = 9.66SVKK428 pKa = 9.88IGNHH432 pKa = 5.86DD433 pKa = 3.86LFAFEE438 pKa = 5.48RR439 pKa = 11.84MISVPGEE446 pKa = 3.99LPLKK450 pKa = 10.37IIYY453 pKa = 7.97EE454 pKa = 4.39CNIFTGCAVYY464 pKa = 10.41SRR466 pKa = 11.84KK467 pKa = 9.62KK468 pKa = 10.42AIMMQAVNSTAQGASTIPGG487 pKa = 3.36
MM1 pKa = 7.77CIFPVSLLSKK11 pKa = 10.7KK12 pKa = 10.4NINCTMVVWPAVLPYY27 pKa = 10.57LEE29 pKa = 4.79AANTVAQGVQTANTIVHH46 pKa = 6.95EE47 pKa = 4.62GQQLAKK53 pKa = 9.47RR54 pKa = 11.84TFGTQISNKK63 pKa = 8.24NKK65 pKa = 9.46KK66 pKa = 9.24RR67 pKa = 11.84KK68 pKa = 9.59LDD70 pKa = 3.46VGHH73 pKa = 7.61ASTQTSGGGKK83 pKa = 9.57YY84 pKa = 9.29YY85 pKa = 9.92QAGYY89 pKa = 9.8VGRR92 pKa = 11.84FRR94 pKa = 11.84GKK96 pKa = 8.34RR97 pKa = 11.84AKK99 pKa = 9.43IVRR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84IPSKK109 pKa = 10.51KK110 pKa = 9.32RR111 pKa = 11.84VAKK114 pKa = 10.23KK115 pKa = 9.59RR116 pKa = 11.84RR117 pKa = 11.84TYY119 pKa = 9.99KK120 pKa = 10.17RR121 pKa = 11.84KK122 pKa = 8.79SRR124 pKa = 11.84VVKK127 pKa = 10.53GEE129 pKa = 3.63NYY131 pKa = 8.67YY132 pKa = 11.27GRR134 pKa = 11.84VGSIATLEE142 pKa = 4.22VTGALSDD149 pKa = 3.78SDD151 pKa = 3.93CVYY154 pKa = 10.42IGHH157 pKa = 6.55SSFAVGQVMRR167 pKa = 11.84MLSVAIMRR175 pKa = 11.84KK176 pKa = 8.09LFKK179 pKa = 9.97MAISWEE185 pKa = 4.06PEE187 pKa = 4.01NISTNIPYY195 pKa = 10.44KK196 pKa = 10.59LVGGAQVSNGYY207 pKa = 7.05TLIVKK212 pKa = 9.99KK213 pKa = 10.71INTDD217 pKa = 3.89DD218 pKa = 3.66NVKK221 pKa = 8.73TQDD224 pKa = 3.64EE225 pKa = 5.16YY226 pKa = 10.9IGPANSHH233 pKa = 6.78NITAPADD240 pKa = 3.55WFRR243 pKa = 11.84TNHH246 pKa = 5.49MEE248 pKa = 5.19AISAQASSNTAVEE261 pKa = 4.3RR262 pKa = 11.84NNRR265 pKa = 11.84LLWLHH270 pKa = 6.17LVDD273 pKa = 3.73VGTGIVRR280 pKa = 11.84AQLDD284 pKa = 4.72LTTLMCDD291 pKa = 3.42VYY293 pKa = 11.33SKK295 pKa = 11.69SNLKK299 pKa = 9.04IQNVTVPSATATEE312 pKa = 4.04ADD314 pKa = 3.44NVNNVPLIGRR324 pKa = 11.84SYY326 pKa = 10.51HH327 pKa = 5.55ISNWQPMTADD337 pKa = 4.37DD338 pKa = 6.29DD339 pKa = 4.06MGPLNRR345 pKa = 11.84LNQDD349 pKa = 3.28TGVLGVEE356 pKa = 4.9IKK358 pKa = 10.65QSATLGSQYY367 pKa = 8.53VTWKK371 pKa = 10.25EE372 pKa = 4.0PPPSKK377 pKa = 10.97AFINCVSSSPEE388 pKa = 3.54RR389 pKa = 11.84MEE391 pKa = 4.65PGTIKK396 pKa = 10.61NHH398 pKa = 4.52VSILRR403 pKa = 11.84RR404 pKa = 11.84SMAFPRR410 pKa = 11.84LMEE413 pKa = 4.33ALAIRR418 pKa = 11.84RR419 pKa = 11.84GNVLNKK425 pKa = 9.66SVKK428 pKa = 9.88IGNHH432 pKa = 5.86DD433 pKa = 3.86LFAFEE438 pKa = 5.48RR439 pKa = 11.84MISVPGEE446 pKa = 3.99LPLKK450 pKa = 10.37IIYY453 pKa = 7.97EE454 pKa = 4.39CNIFTGCAVYY464 pKa = 10.41SRR466 pKa = 11.84KK467 pKa = 9.62KK468 pKa = 10.42AIMMQAVNSTAQGASTIPGG487 pKa = 3.36
Molecular weight: 53.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
789 |
136 |
487 |
263.0 |
28.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.492 ± 0.786 | 1.521 ± 0.324 |
3.802 ± 1.093 | 4.436 ± 0.944 |
2.535 ± 0.356 | 7.858 ± 1.883 |
2.155 ± 0.308 | 4.816 ± 1.938 |
6.084 ± 1.647 | 6.971 ± 0.486 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.788 ± 0.544 | 5.07 ± 1.622 |
6.464 ± 1.988 | 4.689 ± 0.724 |
5.703 ± 1.084 | 7.731 ± 0.172 |
6.337 ± 1.091 | 7.605 ± 1.002 |
2.155 ± 0.85 | 2.788 ± 0.657 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
