
Gramella jeungdoensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Gramella
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
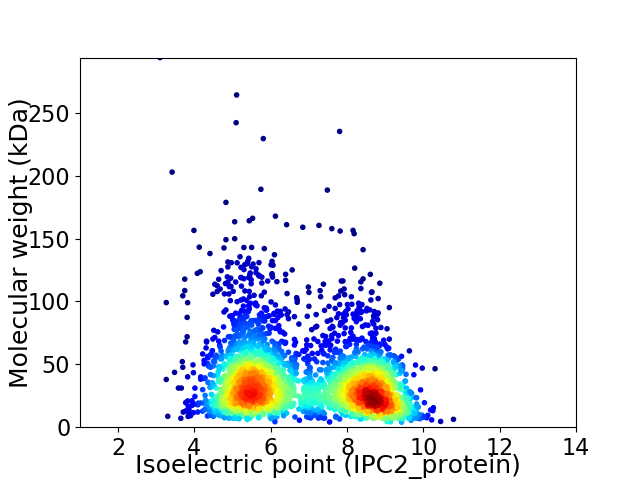
Virtual 2D-PAGE plot for 3093 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y8ATS1|A0A4Y8ATS1_9FLAO Glycoside hydrolase family 95 protein OS=Gramella jeungdoensis OX=708091 GN=E2488_07180 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.15RR3 pKa = 11.84TLLSILLFISFIPFYY18 pKa = 11.02GQIQPFYY25 pKa = 10.93NDD27 pKa = 3.41LDD29 pKa = 3.87FTKK32 pKa = 10.47TEE34 pKa = 3.85NEE36 pKa = 3.92LFIEE40 pKa = 4.56LSNRR44 pKa = 11.84LIDD47 pKa = 3.39THH49 pKa = 5.62TAIPYY54 pKa = 7.79TSSSTDD60 pKa = 2.6VWDD63 pKa = 3.41VLKK66 pKa = 10.86QVDD69 pKa = 4.65EE70 pKa = 5.2DD71 pKa = 4.4PDD73 pKa = 3.65NASNVLLIYY82 pKa = 10.22GFNDD86 pKa = 2.91TDD88 pKa = 4.95GIVDD92 pKa = 3.69TDD94 pKa = 3.44RR95 pKa = 11.84SRR97 pKa = 11.84NKK99 pKa = 10.07NLQDD103 pKa = 3.92SGGGDD108 pKa = 3.2PGRR111 pKa = 11.84WNRR114 pKa = 11.84EE115 pKa = 3.51HH116 pKa = 7.36VFAKK120 pKa = 10.55SLANPSLEE128 pKa = 4.17TGEE131 pKa = 4.89PGPGTDD137 pKa = 3.44VHH139 pKa = 6.2NLKK142 pKa = 10.17PADD145 pKa = 3.67SEE147 pKa = 4.62RR148 pKa = 11.84NSDD151 pKa = 3.07RR152 pKa = 11.84SNRR155 pKa = 11.84KK156 pKa = 8.11FTDD159 pKa = 3.69GSGNSGIVSSNGGWYY174 pKa = 9.72PGDD177 pKa = 3.33EE178 pKa = 4.3WKK180 pKa = 11.21GDD182 pKa = 3.39VARR185 pKa = 11.84IIMYY189 pKa = 8.9MYY191 pKa = 10.58LRR193 pKa = 11.84YY194 pKa = 10.21NGTGAQISQTQCLPINSGIGEE215 pKa = 4.27ALANDD220 pKa = 4.43PNMIDD225 pKa = 6.02LFLNWNIDD233 pKa = 3.66DD234 pKa = 4.24PVSEE238 pKa = 5.39FEE240 pKa = 4.12NQRR243 pKa = 11.84NDD245 pKa = 3.55YY246 pKa = 10.88IEE248 pKa = 4.24TVQGNRR254 pKa = 11.84NPFIDD259 pKa = 3.96NPYY262 pKa = 10.19LATLIWGGVNAEE274 pKa = 4.92DD275 pKa = 4.28KK276 pKa = 10.73WNLNNSSDD284 pKa = 3.96EE285 pKa = 4.17EE286 pKa = 4.78APTSPTNLVALNTTFEE302 pKa = 4.29SLEE305 pKa = 4.32INWDD309 pKa = 3.4AATDD313 pKa = 3.99NIGVIDD319 pKa = 4.33YY320 pKa = 10.63IIYY323 pKa = 10.65LDD325 pKa = 3.77GKK327 pKa = 7.97YY328 pKa = 10.74LKK330 pKa = 8.04TTFSTSSTILEE341 pKa = 4.26LTPNTTYY348 pKa = 11.12NITVKK353 pKa = 10.72ARR355 pKa = 11.84DD356 pKa = 3.52AFSNLSEE363 pKa = 4.21VGTNLSVTTLEE374 pKa = 4.52GPTYY378 pKa = 10.54LINEE382 pKa = 4.45DD383 pKa = 4.6FSNCSNLAFFTYY395 pKa = 10.9NEE397 pKa = 4.1ASNKK401 pKa = 8.97NWEE404 pKa = 4.47CEE406 pKa = 4.06NQFGEE411 pKa = 4.5NNTGSIGINGHH422 pKa = 4.86QQDD425 pKa = 4.53VLSKK429 pKa = 10.17DD430 pKa = 3.17WLITSSPINFDD441 pKa = 3.33EE442 pKa = 4.71NEE444 pKa = 3.89AEE446 pKa = 4.53KK447 pKa = 10.67ISFYY451 pKa = 11.07TDD453 pKa = 2.63AAYY456 pKa = 10.47GNTPLEE462 pKa = 4.06IVYY465 pKa = 10.55SSNYY469 pKa = 9.75DD470 pKa = 3.14GSSNPGQNDD479 pKa = 3.75FTWTPMPNISIPLHH493 pKa = 6.01SNGGATEE500 pKa = 4.49EE501 pKa = 4.52IYY503 pKa = 11.1AFSNVDD509 pKa = 2.81ISSITGTVYY518 pKa = 10.41IAFKK522 pKa = 10.71YY523 pKa = 9.81YY524 pKa = 9.55ATSAPTRR531 pKa = 11.84WTVDD535 pKa = 2.92SFEE538 pKa = 5.81IIADD542 pKa = 3.82NNDD545 pKa = 4.54DD546 pKa = 3.68IDD548 pKa = 5.78GDD550 pKa = 4.36GILNVDD556 pKa = 4.63DD557 pKa = 4.61NCPTTSNTDD566 pKa = 3.19QSDD569 pKa = 3.49TDD571 pKa = 3.54NDD573 pKa = 4.47GVGDD577 pKa = 4.14ACDD580 pKa = 3.54VCPDD584 pKa = 3.83SNDD587 pKa = 4.01TIDD590 pKa = 3.61TDD592 pKa = 4.35GDD594 pKa = 4.49GIPDD598 pKa = 3.94GCDD601 pKa = 2.69VCAEE605 pKa = 4.12SDD607 pKa = 3.74DD608 pKa = 4.86TIDD611 pKa = 3.56TDD613 pKa = 4.6GDD615 pKa = 4.64SIPDD619 pKa = 3.72GCDD622 pKa = 2.88NCPTISNPDD631 pKa = 3.32QADD634 pKa = 3.41TDD636 pKa = 3.88GDD638 pKa = 3.94RR639 pKa = 11.84QGDD642 pKa = 4.01VCDD645 pKa = 3.72STPTGDD651 pKa = 3.38VDD653 pKa = 4.81NDD655 pKa = 3.95GVDD658 pKa = 3.54NAMDD662 pKa = 3.49QCNNTTPGVNVDD674 pKa = 3.33ASGCFTLPANNFTIEE689 pKa = 4.63TISEE693 pKa = 4.23TCPDD697 pKa = 4.03KK698 pKa = 11.67DD699 pKa = 3.44NGQILISAQEE709 pKa = 3.93NYY711 pKa = 9.36TYY713 pKa = 11.2NVTLNGVNATLQNNNLTPGNYY734 pKa = 7.7TVCINVAGEE743 pKa = 4.42SYY745 pKa = 9.16EE746 pKa = 3.73QCYY749 pKa = 10.59DD750 pKa = 3.58VIIAEE755 pKa = 4.45GTTISGKK762 pKa = 9.85VAVTSGKK769 pKa = 10.61ASIQIEE775 pKa = 4.26QGTAPFTVFVNDD787 pKa = 4.68KK788 pKa = 10.47IALEE792 pKa = 4.06SAAPIFTVDD801 pKa = 3.34VKK803 pKa = 11.01HH804 pKa = 6.6GDD806 pKa = 3.59KK807 pKa = 10.67IQVKK811 pKa = 10.41SSVEE815 pKa = 3.86CEE817 pKa = 4.43GIFSKK822 pKa = 10.8SINLFDD828 pKa = 4.89EE829 pKa = 5.06VIAYY833 pKa = 7.79PNPTKK838 pKa = 11.01GNFEE842 pKa = 3.85IATPSNQNNIKK853 pKa = 10.42VEE855 pKa = 4.02IFNMQSQLISAKK867 pKa = 10.65NYY869 pKa = 7.57TLNNGKK875 pKa = 10.15VYY877 pKa = 11.02LNIKK881 pKa = 8.47NQAPGVYY888 pKa = 8.97FAKK891 pKa = 10.62VYY893 pKa = 10.41LDD895 pKa = 3.6KK896 pKa = 11.04PIMLKK901 pKa = 10.1IIKK904 pKa = 9.27EE905 pKa = 4.06
MM1 pKa = 7.44KK2 pKa = 9.15RR3 pKa = 11.84TLLSILLFISFIPFYY18 pKa = 11.02GQIQPFYY25 pKa = 10.93NDD27 pKa = 3.41LDD29 pKa = 3.87FTKK32 pKa = 10.47TEE34 pKa = 3.85NEE36 pKa = 3.92LFIEE40 pKa = 4.56LSNRR44 pKa = 11.84LIDD47 pKa = 3.39THH49 pKa = 5.62TAIPYY54 pKa = 7.79TSSSTDD60 pKa = 2.6VWDD63 pKa = 3.41VLKK66 pKa = 10.86QVDD69 pKa = 4.65EE70 pKa = 5.2DD71 pKa = 4.4PDD73 pKa = 3.65NASNVLLIYY82 pKa = 10.22GFNDD86 pKa = 2.91TDD88 pKa = 4.95GIVDD92 pKa = 3.69TDD94 pKa = 3.44RR95 pKa = 11.84SRR97 pKa = 11.84NKK99 pKa = 10.07NLQDD103 pKa = 3.92SGGGDD108 pKa = 3.2PGRR111 pKa = 11.84WNRR114 pKa = 11.84EE115 pKa = 3.51HH116 pKa = 7.36VFAKK120 pKa = 10.55SLANPSLEE128 pKa = 4.17TGEE131 pKa = 4.89PGPGTDD137 pKa = 3.44VHH139 pKa = 6.2NLKK142 pKa = 10.17PADD145 pKa = 3.67SEE147 pKa = 4.62RR148 pKa = 11.84NSDD151 pKa = 3.07RR152 pKa = 11.84SNRR155 pKa = 11.84KK156 pKa = 8.11FTDD159 pKa = 3.69GSGNSGIVSSNGGWYY174 pKa = 9.72PGDD177 pKa = 3.33EE178 pKa = 4.3WKK180 pKa = 11.21GDD182 pKa = 3.39VARR185 pKa = 11.84IIMYY189 pKa = 8.9MYY191 pKa = 10.58LRR193 pKa = 11.84YY194 pKa = 10.21NGTGAQISQTQCLPINSGIGEE215 pKa = 4.27ALANDD220 pKa = 4.43PNMIDD225 pKa = 6.02LFLNWNIDD233 pKa = 3.66DD234 pKa = 4.24PVSEE238 pKa = 5.39FEE240 pKa = 4.12NQRR243 pKa = 11.84NDD245 pKa = 3.55YY246 pKa = 10.88IEE248 pKa = 4.24TVQGNRR254 pKa = 11.84NPFIDD259 pKa = 3.96NPYY262 pKa = 10.19LATLIWGGVNAEE274 pKa = 4.92DD275 pKa = 4.28KK276 pKa = 10.73WNLNNSSDD284 pKa = 3.96EE285 pKa = 4.17EE286 pKa = 4.78APTSPTNLVALNTTFEE302 pKa = 4.29SLEE305 pKa = 4.32INWDD309 pKa = 3.4AATDD313 pKa = 3.99NIGVIDD319 pKa = 4.33YY320 pKa = 10.63IIYY323 pKa = 10.65LDD325 pKa = 3.77GKK327 pKa = 7.97YY328 pKa = 10.74LKK330 pKa = 8.04TTFSTSSTILEE341 pKa = 4.26LTPNTTYY348 pKa = 11.12NITVKK353 pKa = 10.72ARR355 pKa = 11.84DD356 pKa = 3.52AFSNLSEE363 pKa = 4.21VGTNLSVTTLEE374 pKa = 4.52GPTYY378 pKa = 10.54LINEE382 pKa = 4.45DD383 pKa = 4.6FSNCSNLAFFTYY395 pKa = 10.9NEE397 pKa = 4.1ASNKK401 pKa = 8.97NWEE404 pKa = 4.47CEE406 pKa = 4.06NQFGEE411 pKa = 4.5NNTGSIGINGHH422 pKa = 4.86QQDD425 pKa = 4.53VLSKK429 pKa = 10.17DD430 pKa = 3.17WLITSSPINFDD441 pKa = 3.33EE442 pKa = 4.71NEE444 pKa = 3.89AEE446 pKa = 4.53KK447 pKa = 10.67ISFYY451 pKa = 11.07TDD453 pKa = 2.63AAYY456 pKa = 10.47GNTPLEE462 pKa = 4.06IVYY465 pKa = 10.55SSNYY469 pKa = 9.75DD470 pKa = 3.14GSSNPGQNDD479 pKa = 3.75FTWTPMPNISIPLHH493 pKa = 6.01SNGGATEE500 pKa = 4.49EE501 pKa = 4.52IYY503 pKa = 11.1AFSNVDD509 pKa = 2.81ISSITGTVYY518 pKa = 10.41IAFKK522 pKa = 10.71YY523 pKa = 9.81YY524 pKa = 9.55ATSAPTRR531 pKa = 11.84WTVDD535 pKa = 2.92SFEE538 pKa = 5.81IIADD542 pKa = 3.82NNDD545 pKa = 4.54DD546 pKa = 3.68IDD548 pKa = 5.78GDD550 pKa = 4.36GILNVDD556 pKa = 4.63DD557 pKa = 4.61NCPTTSNTDD566 pKa = 3.19QSDD569 pKa = 3.49TDD571 pKa = 3.54NDD573 pKa = 4.47GVGDD577 pKa = 4.14ACDD580 pKa = 3.54VCPDD584 pKa = 3.83SNDD587 pKa = 4.01TIDD590 pKa = 3.61TDD592 pKa = 4.35GDD594 pKa = 4.49GIPDD598 pKa = 3.94GCDD601 pKa = 2.69VCAEE605 pKa = 4.12SDD607 pKa = 3.74DD608 pKa = 4.86TIDD611 pKa = 3.56TDD613 pKa = 4.6GDD615 pKa = 4.64SIPDD619 pKa = 3.72GCDD622 pKa = 2.88NCPTISNPDD631 pKa = 3.32QADD634 pKa = 3.41TDD636 pKa = 3.88GDD638 pKa = 3.94RR639 pKa = 11.84QGDD642 pKa = 4.01VCDD645 pKa = 3.72STPTGDD651 pKa = 3.38VDD653 pKa = 4.81NDD655 pKa = 3.95GVDD658 pKa = 3.54NAMDD662 pKa = 3.49QCNNTTPGVNVDD674 pKa = 3.33ASGCFTLPANNFTIEE689 pKa = 4.63TISEE693 pKa = 4.23TCPDD697 pKa = 4.03KK698 pKa = 11.67DD699 pKa = 3.44NGQILISAQEE709 pKa = 3.93NYY711 pKa = 9.36TYY713 pKa = 11.2NVTLNGVNATLQNNNLTPGNYY734 pKa = 7.7TVCINVAGEE743 pKa = 4.42SYY745 pKa = 9.16EE746 pKa = 3.73QCYY749 pKa = 10.59DD750 pKa = 3.58VIIAEE755 pKa = 4.45GTTISGKK762 pKa = 9.85VAVTSGKK769 pKa = 10.61ASIQIEE775 pKa = 4.26QGTAPFTVFVNDD787 pKa = 4.68KK788 pKa = 10.47IALEE792 pKa = 4.06SAAPIFTVDD801 pKa = 3.34VKK803 pKa = 11.01HH804 pKa = 6.6GDD806 pKa = 3.59KK807 pKa = 10.67IQVKK811 pKa = 10.41SSVEE815 pKa = 3.86CEE817 pKa = 4.43GIFSKK822 pKa = 10.8SINLFDD828 pKa = 4.89EE829 pKa = 5.06VIAYY833 pKa = 7.79PNPTKK838 pKa = 11.01GNFEE842 pKa = 3.85IATPSNQNNIKK853 pKa = 10.42VEE855 pKa = 4.02IFNMQSQLISAKK867 pKa = 10.65NYY869 pKa = 7.57TLNNGKK875 pKa = 10.15VYY877 pKa = 11.02LNIKK881 pKa = 8.47NQAPGVYY888 pKa = 8.97FAKK891 pKa = 10.62VYY893 pKa = 10.41LDD895 pKa = 3.6KK896 pKa = 11.04PIMLKK901 pKa = 10.1IIKK904 pKa = 9.27EE905 pKa = 4.06
Molecular weight: 98.97 kDa
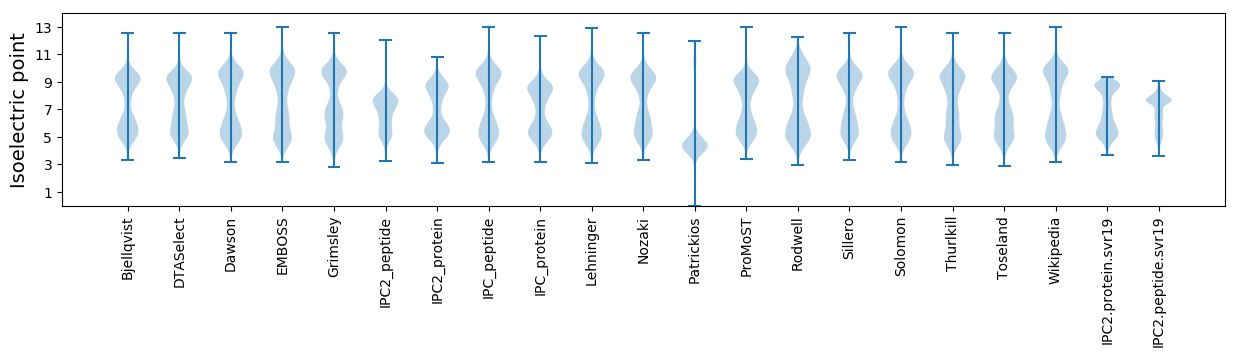
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y8AUH4|A0A4Y8AUH4_9FLAO L-fucose:H+ symporter permease OS=Gramella jeungdoensis OX=708091 GN=fucP PE=4 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.24RR22 pKa = 11.84MASATGRR29 pKa = 11.84KK30 pKa = 8.64VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.02GRR40 pKa = 11.84NRR42 pKa = 11.84VSVSSDD48 pKa = 3.1PRR50 pKa = 11.84PKK52 pKa = 10.39KK53 pKa = 10.52
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.24RR22 pKa = 11.84MASATGRR29 pKa = 11.84KK30 pKa = 8.64VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.02GRR40 pKa = 11.84NRR42 pKa = 11.84VSVSSDD48 pKa = 3.1PRR50 pKa = 11.84PKK52 pKa = 10.39KK53 pKa = 10.52
Molecular weight: 6.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1079700 |
34 |
2820 |
349.1 |
39.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.094 ± 0.039 | 0.74 ± 0.014 |
5.171 ± 0.036 | 6.654 ± 0.04 |
5.363 ± 0.034 | 6.221 ± 0.043 |
1.667 ± 0.019 | 8.696 ± 0.055 |
8.506 ± 0.057 | 9.265 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.056 ± 0.026 | 6.733 ± 0.049 |
3.243 ± 0.021 | 3.1 ± 0.024 |
3.068 ± 0.03 | 6.444 ± 0.036 |
5.841 ± 0.046 | 6.051 ± 0.035 |
1.053 ± 0.017 | 4.035 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
