
Campoletis sonorensis ichnovirus (strain Texas A&M) (CsIV)
Taxonomy: Viruses; Polydnaviridae; Ichnovirus; Campoletis sonorensis ichnovirus
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
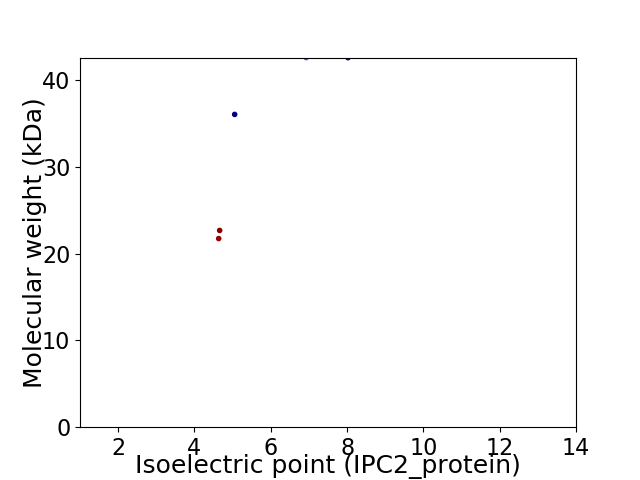
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
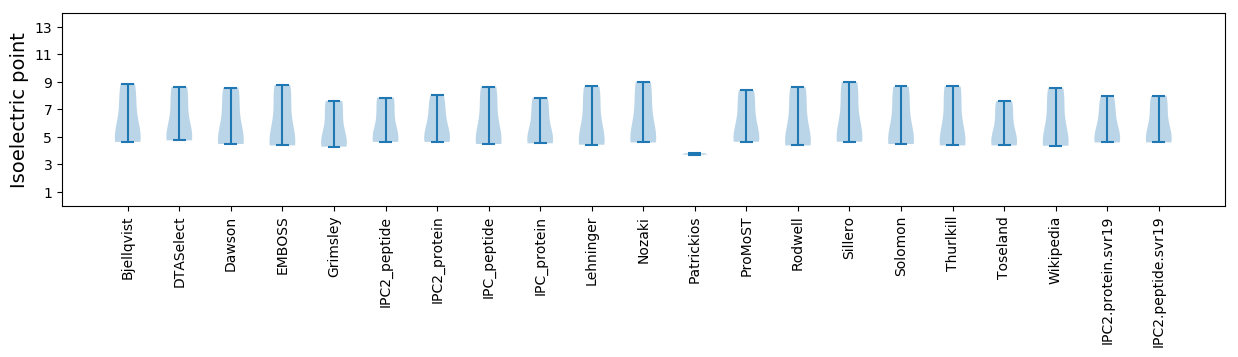
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q77ZH4|Q77ZH4_CSIVT Cysteine-rich protein OS=Campoletis sonorensis ichnovirus (strain Texas A&M) OX=654920 GN=WHv1.0 PE=4 SV=1
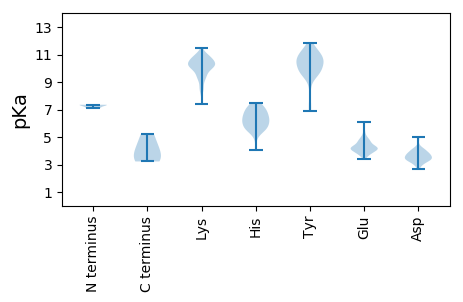
MM1 pKa = 7.19KK2 pKa = 10.53VLWFLLGAAVMVQARR17 pKa = 11.84PDD19 pKa = 3.52MVSGEE24 pKa = 4.33KK25 pKa = 10.51SGDD28 pKa = 3.21WASEE32 pKa = 4.16TEE34 pKa = 4.77FIVTEE39 pKa = 4.61DD40 pKa = 4.07GIKK43 pKa = 10.53PNPNYY48 pKa = 10.23VKK50 pKa = 10.31QDD52 pKa = 3.63GVSSPPVSGWTEE64 pKa = 3.67DD65 pKa = 3.4RR66 pKa = 11.84EE67 pKa = 4.32FAVTSSTAGPEE78 pKa = 4.21PKK80 pKa = 10.27SSVINIDD87 pKa = 3.07VTKK90 pKa = 10.35PITAEE95 pKa = 3.64LTINFHH101 pKa = 5.55QVQDD105 pKa = 4.13PAPKK109 pKa = 9.93PEE111 pKa = 4.62PSCIDD116 pKa = 2.66NWKK119 pKa = 8.82YY120 pKa = 10.7CRR122 pKa = 11.84GINKK126 pKa = 8.17PCCGQQLMEE135 pKa = 6.03DD136 pKa = 3.8GTLGPKK142 pKa = 10.18HH143 pKa = 5.78FVCFEE148 pKa = 4.62LGQGICTPLSQIKK161 pKa = 10.24NADD164 pKa = 3.8LFAKK168 pKa = 10.56LADD171 pKa = 3.7QLNDD175 pKa = 3.14TNYY178 pKa = 11.22AEE180 pKa = 6.1LEE182 pKa = 4.09SQYY185 pKa = 9.59WKK187 pKa = 10.55SVSSPEE193 pKa = 4.18SSSTFF198 pKa = 3.22
MM1 pKa = 7.19KK2 pKa = 10.53VLWFLLGAAVMVQARR17 pKa = 11.84PDD19 pKa = 3.52MVSGEE24 pKa = 4.33KK25 pKa = 10.51SGDD28 pKa = 3.21WASEE32 pKa = 4.16TEE34 pKa = 4.77FIVTEE39 pKa = 4.61DD40 pKa = 4.07GIKK43 pKa = 10.53PNPNYY48 pKa = 10.23VKK50 pKa = 10.31QDD52 pKa = 3.63GVSSPPVSGWTEE64 pKa = 3.67DD65 pKa = 3.4RR66 pKa = 11.84EE67 pKa = 4.32FAVTSSTAGPEE78 pKa = 4.21PKK80 pKa = 10.27SSVINIDD87 pKa = 3.07VTKK90 pKa = 10.35PITAEE95 pKa = 3.64LTINFHH101 pKa = 5.55QVQDD105 pKa = 4.13PAPKK109 pKa = 9.93PEE111 pKa = 4.62PSCIDD116 pKa = 2.66NWKK119 pKa = 8.82YY120 pKa = 10.7CRR122 pKa = 11.84GINKK126 pKa = 8.17PCCGQQLMEE135 pKa = 6.03DD136 pKa = 3.8GTLGPKK142 pKa = 10.18HH143 pKa = 5.78FVCFEE148 pKa = 4.62LGQGICTPLSQIKK161 pKa = 10.24NADD164 pKa = 3.8LFAKK168 pKa = 10.56LADD171 pKa = 3.7QLNDD175 pKa = 3.14TNYY178 pKa = 11.22AEE180 pKa = 6.1LEE182 pKa = 4.09SQYY185 pKa = 9.59WKK187 pKa = 10.55SVSSPEE193 pKa = 4.18SSSTFF198 pKa = 3.22
Molecular weight: 21.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8JV09|Q8JV09_CSIVT Innexin-like protein 2 OS=Campoletis sonorensis ichnovirus (strain Texas A&M) OX=654920 PE=3 SV=1
MM1 pKa = 7.35LKK3 pKa = 9.87IFRR6 pKa = 11.84TLRR9 pKa = 11.84GLLKK13 pKa = 10.47VHH15 pKa = 6.97VISIDD20 pKa = 3.33NNFFILHH27 pKa = 5.04YY28 pKa = 9.75KK29 pKa = 7.43ITVVILLALAMLVTSQQFFKK49 pKa = 11.49NPMEE53 pKa = 5.0CNFSDD58 pKa = 4.13LPLGSSHH65 pKa = 6.23YY66 pKa = 9.8CYY68 pKa = 10.98VHH70 pKa = 6.29ATFLEE75 pKa = 4.34QQQITHH81 pKa = 6.66HH82 pKa = 6.26VPPQRR87 pKa = 11.84LPGGNISGEE96 pKa = 4.16TGEE99 pKa = 4.81KK100 pKa = 10.14EE101 pKa = 3.77FRR103 pKa = 11.84FYY105 pKa = 11.36NYY107 pKa = 10.47YY108 pKa = 9.19EE109 pKa = 4.06WVYY112 pKa = 9.98LTLAVQAILFYY123 pKa = 10.52VPHH126 pKa = 6.68YY127 pKa = 9.73IWKK130 pKa = 8.48AWEE133 pKa = 4.35GGKK136 pKa = 9.11MKK138 pKa = 10.05MLAVEE143 pKa = 4.73FASPVLSEE151 pKa = 5.19DD152 pKa = 3.77FIEE155 pKa = 4.4NKK157 pKa = 9.32MIPVVEE163 pKa = 4.23YY164 pKa = 10.56FCTTLHH170 pKa = 5.79SHH172 pKa = 6.08NAYY175 pKa = 9.39AYY177 pKa = 9.65KK178 pKa = 10.79YY179 pKa = 6.86FTCEE183 pKa = 3.49FLNLVNVVGQILFLKK198 pKa = 10.19IFLGEE203 pKa = 3.92EE204 pKa = 4.01FASFGIDD211 pKa = 3.71VITFDD216 pKa = 3.92HH217 pKa = 6.23RR218 pKa = 11.84QEE220 pKa = 4.14KK221 pKa = 10.29SMKK224 pKa = 10.31NPIDD228 pKa = 3.39RR229 pKa = 11.84LFPIVTRR236 pKa = 11.84CSYY239 pKa = 10.81HH240 pKa = 6.96KK241 pKa = 10.02YY242 pKa = 9.42GPSGKK247 pKa = 9.51VEE249 pKa = 3.84NWEE252 pKa = 4.19GLCLLPEE259 pKa = 4.12NSLNGKK265 pKa = 9.48IYY267 pKa = 10.37IFMWFWFHH275 pKa = 6.48MLTAISSVVVIYY287 pKa = 10.34RR288 pKa = 11.84IVTLCSPSVRR298 pKa = 11.84LYY300 pKa = 10.62RR301 pKa = 11.84FKK303 pKa = 10.68PLSGLIRR310 pKa = 11.84SEE312 pKa = 4.83DD313 pKa = 3.33IAIVFPKK320 pKa = 10.35LNVGIGFCCVGCNGISTRR338 pKa = 11.84LHH340 pKa = 5.44TKK342 pKa = 10.18RR343 pKa = 11.84SFLALQSTLARR354 pKa = 11.84PLQMLKK360 pKa = 10.74VIADD364 pKa = 3.52VKK366 pKa = 10.8LALL369 pKa = 4.36
MM1 pKa = 7.35LKK3 pKa = 9.87IFRR6 pKa = 11.84TLRR9 pKa = 11.84GLLKK13 pKa = 10.47VHH15 pKa = 6.97VISIDD20 pKa = 3.33NNFFILHH27 pKa = 5.04YY28 pKa = 9.75KK29 pKa = 7.43ITVVILLALAMLVTSQQFFKK49 pKa = 11.49NPMEE53 pKa = 5.0CNFSDD58 pKa = 4.13LPLGSSHH65 pKa = 6.23YY66 pKa = 9.8CYY68 pKa = 10.98VHH70 pKa = 6.29ATFLEE75 pKa = 4.34QQQITHH81 pKa = 6.66HH82 pKa = 6.26VPPQRR87 pKa = 11.84LPGGNISGEE96 pKa = 4.16TGEE99 pKa = 4.81KK100 pKa = 10.14EE101 pKa = 3.77FRR103 pKa = 11.84FYY105 pKa = 11.36NYY107 pKa = 10.47YY108 pKa = 9.19EE109 pKa = 4.06WVYY112 pKa = 9.98LTLAVQAILFYY123 pKa = 10.52VPHH126 pKa = 6.68YY127 pKa = 9.73IWKK130 pKa = 8.48AWEE133 pKa = 4.35GGKK136 pKa = 9.11MKK138 pKa = 10.05MLAVEE143 pKa = 4.73FASPVLSEE151 pKa = 5.19DD152 pKa = 3.77FIEE155 pKa = 4.4NKK157 pKa = 9.32MIPVVEE163 pKa = 4.23YY164 pKa = 10.56FCTTLHH170 pKa = 5.79SHH172 pKa = 6.08NAYY175 pKa = 9.39AYY177 pKa = 9.65KK178 pKa = 10.79YY179 pKa = 6.86FTCEE183 pKa = 3.49FLNLVNVVGQILFLKK198 pKa = 10.19IFLGEE203 pKa = 3.92EE204 pKa = 4.01FASFGIDD211 pKa = 3.71VITFDD216 pKa = 3.92HH217 pKa = 6.23RR218 pKa = 11.84QEE220 pKa = 4.14KK221 pKa = 10.29SMKK224 pKa = 10.31NPIDD228 pKa = 3.39RR229 pKa = 11.84LFPIVTRR236 pKa = 11.84CSYY239 pKa = 10.81HH240 pKa = 6.96KK241 pKa = 10.02YY242 pKa = 9.42GPSGKK247 pKa = 9.51VEE249 pKa = 3.84NWEE252 pKa = 4.19GLCLLPEE259 pKa = 4.12NSLNGKK265 pKa = 9.48IYY267 pKa = 10.37IFMWFWFHH275 pKa = 6.48MLTAISSVVVIYY287 pKa = 10.34RR288 pKa = 11.84IVTLCSPSVRR298 pKa = 11.84LYY300 pKa = 10.62RR301 pKa = 11.84FKK303 pKa = 10.68PLSGLIRR310 pKa = 11.84SEE312 pKa = 4.83DD313 pKa = 3.33IAIVFPKK320 pKa = 10.35LNVGIGFCCVGCNGISTRR338 pKa = 11.84LHH340 pKa = 5.44TKK342 pKa = 10.18RR343 pKa = 11.84SFLALQSTLARR354 pKa = 11.84PLQMLKK360 pKa = 10.74VIADD364 pKa = 3.52VKK366 pKa = 10.8LALL369 pKa = 4.36
Molecular weight: 42.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1457 |
198 |
369 |
291.4 |
33.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.873 ± 0.271 | 3.02 ± 0.479 |
4.53 ± 0.761 | 7.001 ± 1.119 |
5.971 ± 0.761 | 5.559 ± 0.423 |
2.471 ± 0.51 | 6.452 ± 0.561 |
6.04 ± 0.595 | 9.54 ± 1.003 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.402 ± 0.381 | 4.736 ± 0.283 |
5.903 ± 0.717 | 3.089 ± 0.356 |
3.569 ± 0.546 | 7.344 ± 0.59 |
5.353 ± 0.213 | 6.726 ± 0.413 |
1.647 ± 0.163 | 3.775 ± 0.618 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
