
Bahia Grande virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Barhavirus; Bahia barhavirus
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
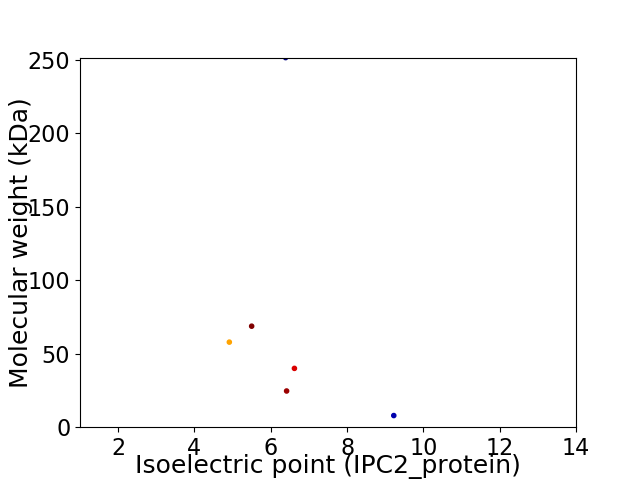
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D3R266|A0A0D3R266_9RHAB GDP polyribonucleotidyltransferase OS=Bahia Grande virus OX=932699 PE=4 SV=1
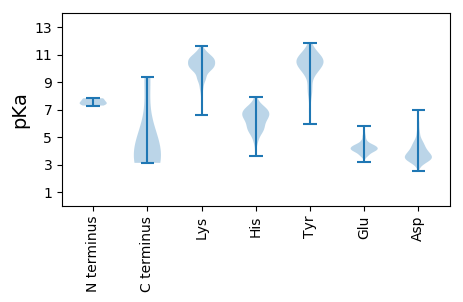
MM1 pKa = 7.52AAAILPVSRR10 pKa = 11.84NMPVRR15 pKa = 11.84EE16 pKa = 3.85RR17 pKa = 11.84TVAGSVTAPPVQYY30 pKa = 10.42PSTWFQAHH38 pKa = 7.22AGQKK42 pKa = 10.03VSITIYY48 pKa = 10.13QNTNARR54 pKa = 11.84QAFSRR59 pKa = 11.84ITQLRR64 pKa = 11.84NNGQWDD70 pKa = 4.05DD71 pKa = 3.77KK72 pKa = 11.49LIATFMKK79 pKa = 10.7GVLDD83 pKa = 5.18EE84 pKa = 4.2NAEE87 pKa = 4.23WFQSPPLIEE96 pKa = 4.27DD97 pKa = 4.1WIVNEE102 pKa = 3.94AVIGRR107 pKa = 11.84VDD109 pKa = 4.31DD110 pKa = 4.31VVAPTALAQWEE121 pKa = 4.42EE122 pKa = 4.26VEE124 pKa = 5.18RR125 pKa = 11.84PQNMDD130 pKa = 3.53PVPNEE135 pKa = 3.7EE136 pKa = 4.61GEE138 pKa = 4.22LGTRR142 pKa = 11.84RR143 pKa = 11.84SFFLALITIYY153 pKa = 10.51RR154 pKa = 11.84QVLTRR159 pKa = 11.84TINVDD164 pKa = 3.05YY165 pKa = 9.93GQEE168 pKa = 3.69VSRR171 pKa = 11.84RR172 pKa = 11.84IIDD175 pKa = 3.59NFKK178 pKa = 9.93EE179 pKa = 4.17QPLGMSQDD187 pKa = 4.91DD188 pKa = 3.5INEE191 pKa = 3.95IQGYY195 pKa = 6.75EE196 pKa = 4.03SKK198 pKa = 10.88EE199 pKa = 3.91RR200 pKa = 11.84LTTNYY205 pKa = 9.99VKK207 pKa = 10.35ILCILDD213 pKa = 3.17MFFNKK218 pKa = 9.61FQTHH222 pKa = 6.87DD223 pKa = 3.18KK224 pKa = 8.98STIRR228 pKa = 11.84IATLPTRR235 pKa = 11.84YY236 pKa = 9.38RR237 pKa = 11.84GCAAFTSYY245 pKa = 11.65GEE247 pKa = 3.81LAIRR251 pKa = 11.84LGIEE255 pKa = 4.31PIKK258 pKa = 10.67LPSLILTVAVAKK270 pKa = 10.81DD271 pKa = 3.39FDD273 pKa = 4.72KK274 pKa = 11.34INVNGEE280 pKa = 3.64QAEE283 pKa = 4.12QLDD286 pKa = 4.81GYY288 pKa = 10.81FPYY291 pKa = 10.44QLEE294 pKa = 4.24LGLVKK299 pKa = 10.45KK300 pKa = 10.36SAYY303 pKa = 9.96SAGNCPSLYY312 pKa = 10.7LWMHH316 pKa = 6.72TIGTMLHH323 pKa = 4.69QQRR326 pKa = 11.84SYY328 pKa = 10.61RR329 pKa = 11.84ANVPKK334 pKa = 10.53NVPDD338 pKa = 3.35QMGTINSAIAVAMQFVAGGEE358 pKa = 3.93FSMQFVGDD366 pKa = 3.84ARR368 pKa = 11.84VQEE371 pKa = 4.75AMRR374 pKa = 11.84EE375 pKa = 4.18MQTAEE380 pKa = 4.04AEE382 pKa = 4.25LNEE385 pKa = 4.27LRR387 pKa = 11.84MAQARR392 pKa = 11.84EE393 pKa = 3.63MRR395 pKa = 11.84AAARR399 pKa = 11.84GDD401 pKa = 3.39EE402 pKa = 4.29DD403 pKa = 4.77EE404 pKa = 5.13EE405 pKa = 4.97GSEE408 pKa = 5.74DD409 pKa = 5.13GLDD412 pKa = 4.16DD413 pKa = 4.55EE414 pKa = 5.8NDD416 pKa = 3.99GEE418 pKa = 5.46GDD420 pKa = 3.81DD421 pKa = 4.47EE422 pKa = 5.01LPAEE426 pKa = 4.4IEE428 pKa = 4.18QNPEE432 pKa = 3.23YY433 pKa = 10.57LNRR436 pKa = 11.84VNRR439 pKa = 11.84IRR441 pKa = 11.84EE442 pKa = 4.15LQEE445 pKa = 4.33NLQQYY450 pKa = 8.86NATVQQHH457 pKa = 4.58TNAVEE462 pKa = 4.03KK463 pKa = 10.67AALRR467 pKa = 11.84ALAYY471 pKa = 9.99LQEE474 pKa = 4.51NGGIADD480 pKa = 3.62KK481 pKa = 10.89DD482 pKa = 3.71KK483 pKa = 11.09RR484 pKa = 11.84DD485 pKa = 3.23LGIRR489 pKa = 11.84FRR491 pKa = 11.84RR492 pKa = 11.84FADD495 pKa = 3.51EE496 pKa = 4.09AEE498 pKa = 4.09GRR500 pKa = 11.84VGKK503 pKa = 10.23LLASLFPAPRR513 pKa = 4.8
MM1 pKa = 7.52AAAILPVSRR10 pKa = 11.84NMPVRR15 pKa = 11.84EE16 pKa = 3.85RR17 pKa = 11.84TVAGSVTAPPVQYY30 pKa = 10.42PSTWFQAHH38 pKa = 7.22AGQKK42 pKa = 10.03VSITIYY48 pKa = 10.13QNTNARR54 pKa = 11.84QAFSRR59 pKa = 11.84ITQLRR64 pKa = 11.84NNGQWDD70 pKa = 4.05DD71 pKa = 3.77KK72 pKa = 11.49LIATFMKK79 pKa = 10.7GVLDD83 pKa = 5.18EE84 pKa = 4.2NAEE87 pKa = 4.23WFQSPPLIEE96 pKa = 4.27DD97 pKa = 4.1WIVNEE102 pKa = 3.94AVIGRR107 pKa = 11.84VDD109 pKa = 4.31DD110 pKa = 4.31VVAPTALAQWEE121 pKa = 4.42EE122 pKa = 4.26VEE124 pKa = 5.18RR125 pKa = 11.84PQNMDD130 pKa = 3.53PVPNEE135 pKa = 3.7EE136 pKa = 4.61GEE138 pKa = 4.22LGTRR142 pKa = 11.84RR143 pKa = 11.84SFFLALITIYY153 pKa = 10.51RR154 pKa = 11.84QVLTRR159 pKa = 11.84TINVDD164 pKa = 3.05YY165 pKa = 9.93GQEE168 pKa = 3.69VSRR171 pKa = 11.84RR172 pKa = 11.84IIDD175 pKa = 3.59NFKK178 pKa = 9.93EE179 pKa = 4.17QPLGMSQDD187 pKa = 4.91DD188 pKa = 3.5INEE191 pKa = 3.95IQGYY195 pKa = 6.75EE196 pKa = 4.03SKK198 pKa = 10.88EE199 pKa = 3.91RR200 pKa = 11.84LTTNYY205 pKa = 9.99VKK207 pKa = 10.35ILCILDD213 pKa = 3.17MFFNKK218 pKa = 9.61FQTHH222 pKa = 6.87DD223 pKa = 3.18KK224 pKa = 8.98STIRR228 pKa = 11.84IATLPTRR235 pKa = 11.84YY236 pKa = 9.38RR237 pKa = 11.84GCAAFTSYY245 pKa = 11.65GEE247 pKa = 3.81LAIRR251 pKa = 11.84LGIEE255 pKa = 4.31PIKK258 pKa = 10.67LPSLILTVAVAKK270 pKa = 10.81DD271 pKa = 3.39FDD273 pKa = 4.72KK274 pKa = 11.34INVNGEE280 pKa = 3.64QAEE283 pKa = 4.12QLDD286 pKa = 4.81GYY288 pKa = 10.81FPYY291 pKa = 10.44QLEE294 pKa = 4.24LGLVKK299 pKa = 10.45KK300 pKa = 10.36SAYY303 pKa = 9.96SAGNCPSLYY312 pKa = 10.7LWMHH316 pKa = 6.72TIGTMLHH323 pKa = 4.69QQRR326 pKa = 11.84SYY328 pKa = 10.61RR329 pKa = 11.84ANVPKK334 pKa = 10.53NVPDD338 pKa = 3.35QMGTINSAIAVAMQFVAGGEE358 pKa = 3.93FSMQFVGDD366 pKa = 3.84ARR368 pKa = 11.84VQEE371 pKa = 4.75AMRR374 pKa = 11.84EE375 pKa = 4.18MQTAEE380 pKa = 4.04AEE382 pKa = 4.25LNEE385 pKa = 4.27LRR387 pKa = 11.84MAQARR392 pKa = 11.84EE393 pKa = 3.63MRR395 pKa = 11.84AAARR399 pKa = 11.84GDD401 pKa = 3.39EE402 pKa = 4.29DD403 pKa = 4.77EE404 pKa = 5.13EE405 pKa = 4.97GSEE408 pKa = 5.74DD409 pKa = 5.13GLDD412 pKa = 4.16DD413 pKa = 4.55EE414 pKa = 5.8NDD416 pKa = 3.99GEE418 pKa = 5.46GDD420 pKa = 3.81DD421 pKa = 4.47EE422 pKa = 5.01LPAEE426 pKa = 4.4IEE428 pKa = 4.18QNPEE432 pKa = 3.23YY433 pKa = 10.57LNRR436 pKa = 11.84VNRR439 pKa = 11.84IRR441 pKa = 11.84EE442 pKa = 4.15LQEE445 pKa = 4.33NLQQYY450 pKa = 8.86NATVQQHH457 pKa = 4.58TNAVEE462 pKa = 4.03KK463 pKa = 10.67AALRR467 pKa = 11.84ALAYY471 pKa = 9.99LQEE474 pKa = 4.51NGGIADD480 pKa = 3.62KK481 pKa = 10.89DD482 pKa = 3.71KK483 pKa = 11.09RR484 pKa = 11.84DD485 pKa = 3.23LGIRR489 pKa = 11.84FRR491 pKa = 11.84RR492 pKa = 11.84FADD495 pKa = 3.51EE496 pKa = 4.09AEE498 pKa = 4.09GRR500 pKa = 11.84VGKK503 pKa = 10.23LLASLFPAPRR513 pKa = 4.8
Molecular weight: 57.85 kDa
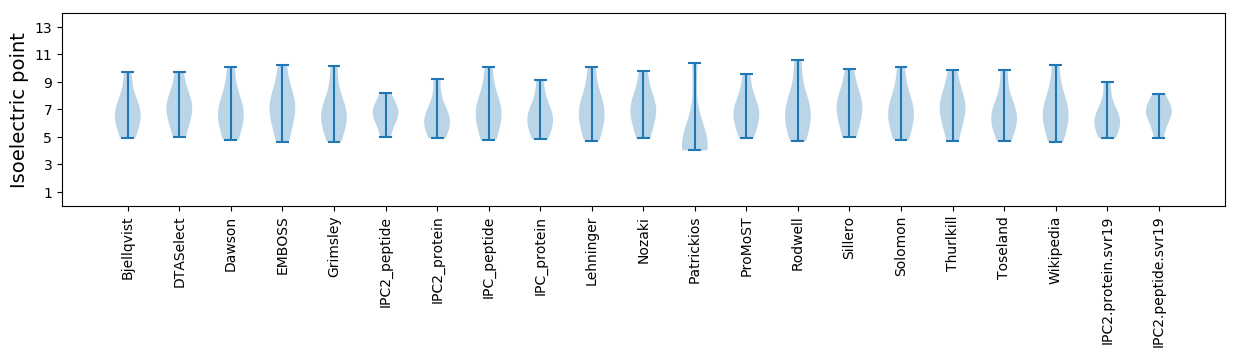
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D3R273|A0A0D3R273_9RHAB Uncharacterized protein OS=Bahia Grande virus OX=932699 PE=4 SV=1
MM1 pKa = 7.55PQHH4 pKa = 5.57QTLSQDD10 pKa = 3.63PPQQPIVFSTVKK22 pKa = 9.99KK23 pKa = 9.51VGYY26 pKa = 9.65IILFSIPLIVIIVFFVLLDD45 pKa = 3.89RR46 pKa = 11.84KK47 pKa = 9.28HH48 pKa = 6.35LKK50 pKa = 10.22YY51 pKa = 10.48IPLKK55 pKa = 10.35AWNTSIITIKK65 pKa = 10.48EE66 pKa = 3.92PYY68 pKa = 9.35
MM1 pKa = 7.55PQHH4 pKa = 5.57QTLSQDD10 pKa = 3.63PPQQPIVFSTVKK22 pKa = 9.99KK23 pKa = 9.51VGYY26 pKa = 9.65IILFSIPLIVIIVFFVLLDD45 pKa = 3.89RR46 pKa = 11.84KK47 pKa = 9.28HH48 pKa = 6.35LKK50 pKa = 10.22YY51 pKa = 10.48IPLKK55 pKa = 10.35AWNTSIITIKK65 pKa = 10.48EE66 pKa = 3.92PYY68 pKa = 9.35
Molecular weight: 7.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3920 |
68 |
2175 |
653.3 |
75.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.49 ± 0.929 | 1.327 ± 0.318 |
5.689 ± 0.173 | 7.781 ± 0.349 |
4.362 ± 0.355 | 5.51 ± 0.295 |
2.143 ± 0.221 | 7.628 ± 0.356 |
7.526 ± 0.955 | 9.464 ± 0.767 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.372 ± 0.104 | 5.638 ± 0.314 |
4.056 ± 0.382 | 3.546 ± 0.675 |
5.051 ± 0.505 | 6.964 ± 0.489 |
5.791 ± 0.168 | 5.23 ± 0.288 |
1.48 ± 0.289 | 3.954 ± 0.414 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
