
Halobacterium jilantaiense
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Halobacteriaceae; Halobacterium
Average proteome isoelectric point is 4.8
Get precalculated fractions of proteins
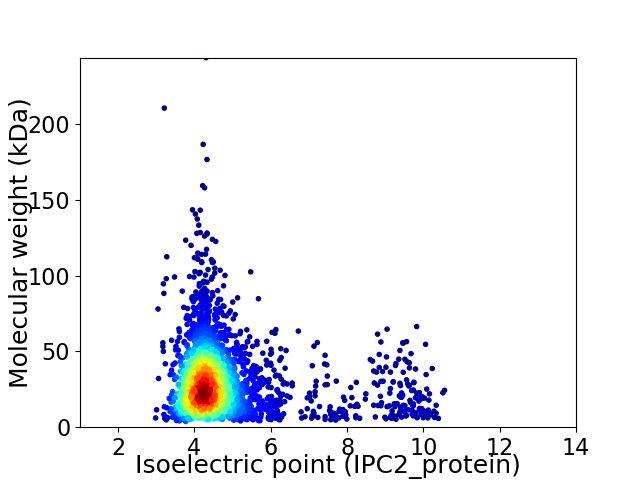
Virtual 2D-PAGE plot for 3054 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
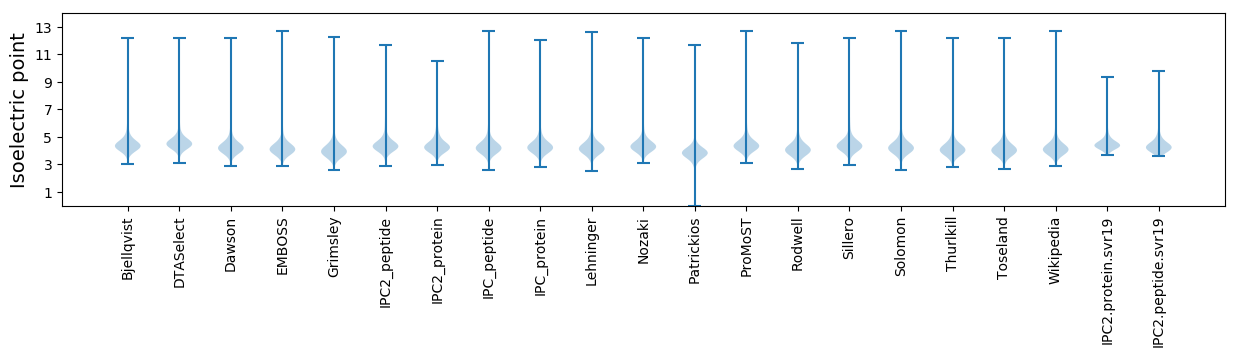
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I0PJ56|A0A1I0PJ56_9EURY Uncharacterized protein OS=Halobacterium jilantaiense OX=355548 GN=SAMN04487945_1731 PE=4 SV=1
MM1 pKa = 7.82ADD3 pKa = 4.17DD4 pKa = 3.65NTLTRR9 pKa = 11.84RR10 pKa = 11.84SFLTATGAAAGSVAFVGAGAATPGKK35 pKa = 10.54GRR37 pKa = 11.84GNGRR41 pKa = 11.84GKK43 pKa = 8.87GTEE46 pKa = 3.53RR47 pKa = 11.84RR48 pKa = 11.84EE49 pKa = 4.18YY50 pKa = 10.28IVGVSASEE58 pKa = 4.09DD59 pKa = 3.48DD60 pKa = 3.39PRR62 pKa = 11.84GYY64 pKa = 10.01VRR66 pKa = 11.84KK67 pKa = 9.19HH68 pKa = 5.47LSGKK72 pKa = 8.81EE73 pKa = 3.87EE74 pKa = 4.77PISTNEE80 pKa = 3.82RR81 pKa = 11.84LGTTVVGLPEE91 pKa = 4.01NAAEE95 pKa = 3.97QAHH98 pKa = 5.25QNFLGRR104 pKa = 11.84LEE106 pKa = 4.38AADD109 pKa = 3.46GVKK112 pKa = 10.3YY113 pKa = 10.69VEE115 pKa = 4.55QNVDD119 pKa = 3.23LQAQVAPNDD128 pKa = 3.24PRR130 pKa = 11.84YY131 pKa = 9.58RR132 pKa = 11.84DD133 pKa = 3.4QYY135 pKa = 11.93ADD137 pKa = 3.45QQVNAPEE144 pKa = 4.2AWDD147 pKa = 3.55ATFGDD152 pKa = 3.83SGVTVAVVDD161 pKa = 3.52QGVKK165 pKa = 10.09YY166 pKa = 10.07DD167 pKa = 4.85HH168 pKa = 7.72PDD170 pKa = 3.02LDD172 pKa = 4.3GNMDD176 pKa = 3.71GSVSNSGYY184 pKa = 11.13DD185 pKa = 4.3FVDD188 pKa = 5.06DD189 pKa = 6.14DD190 pKa = 5.45DD191 pKa = 6.29DD192 pKa = 5.04PYY194 pKa = 11.53PDD196 pKa = 4.25ALSDD200 pKa = 3.91EE201 pKa = 4.56YY202 pKa = 11.15HH203 pKa = 5.64GTHH206 pKa = 6.07VGGIAAAEE214 pKa = 4.02VDD216 pKa = 3.5NGEE219 pKa = 4.19GVTGIGNSSLLSGRR233 pKa = 11.84ALSEE237 pKa = 3.94QGSGSTADD245 pKa = 3.35IADD248 pKa = 3.88AVTWAADD255 pKa = 3.29QGADD259 pKa = 4.73VINLSLGGGGYY270 pKa = 8.89TDD272 pKa = 3.24TMKK275 pKa = 10.92NAVSYY280 pKa = 8.31ATNNGALVVAAAGNDD295 pKa = 3.82GQGSVSYY302 pKa = 9.27PAAYY306 pKa = 9.87SEE308 pKa = 4.45CLAVSALDD316 pKa = 3.77PDD318 pKa = 3.94EE319 pKa = 4.65TLASYY324 pKa = 11.31SNYY327 pKa = 10.19GGNIEE332 pKa = 4.42LAAPGTNVLSTWTDD346 pKa = 3.17DD347 pKa = 4.0AYY349 pKa = 11.12DD350 pKa = 4.39SISGTSMATPVVAGVAGLALAQWDD374 pKa = 4.01LTNAEE379 pKa = 4.73LRR381 pKa = 11.84NHH383 pKa = 7.19LKK385 pKa = 8.53NTAVNVGLSSDD396 pKa = 3.43KK397 pKa = 10.86QGSGRR402 pKa = 11.84VDD404 pKa = 3.09AGNAITTEE412 pKa = 4.31PGSGGGGGGGGGGSTSTTISGSLSSYY438 pKa = 10.99ADD440 pKa = 3.85SDD442 pKa = 3.81CSTYY446 pKa = 10.13TFEE449 pKa = 4.91YY450 pKa = 10.23DD451 pKa = 3.33SPSQIVIEE459 pKa = 4.16LSGPSDD465 pKa = 4.11ADD467 pKa = 3.05FDD469 pKa = 5.2LYY471 pKa = 11.65ANTGVGACPTTDD483 pKa = 3.35DD484 pKa = 3.72YY485 pKa = 11.81DD486 pKa = 3.87YY487 pKa = 11.49RR488 pKa = 11.84SYY490 pKa = 8.17TTNSQEE496 pKa = 4.43TITIDD501 pKa = 4.4SPDD504 pKa = 3.67TSTDD508 pKa = 2.99LYY510 pKa = 11.42ALVDD514 pKa = 4.1SYY516 pKa = 11.78SGSGDD521 pKa = 3.28FTVTFTEE528 pKa = 4.51YY529 pKa = 10.23QQ530 pKa = 3.16
MM1 pKa = 7.82ADD3 pKa = 4.17DD4 pKa = 3.65NTLTRR9 pKa = 11.84RR10 pKa = 11.84SFLTATGAAAGSVAFVGAGAATPGKK35 pKa = 10.54GRR37 pKa = 11.84GNGRR41 pKa = 11.84GKK43 pKa = 8.87GTEE46 pKa = 3.53RR47 pKa = 11.84RR48 pKa = 11.84EE49 pKa = 4.18YY50 pKa = 10.28IVGVSASEE58 pKa = 4.09DD59 pKa = 3.48DD60 pKa = 3.39PRR62 pKa = 11.84GYY64 pKa = 10.01VRR66 pKa = 11.84KK67 pKa = 9.19HH68 pKa = 5.47LSGKK72 pKa = 8.81EE73 pKa = 3.87EE74 pKa = 4.77PISTNEE80 pKa = 3.82RR81 pKa = 11.84LGTTVVGLPEE91 pKa = 4.01NAAEE95 pKa = 3.97QAHH98 pKa = 5.25QNFLGRR104 pKa = 11.84LEE106 pKa = 4.38AADD109 pKa = 3.46GVKK112 pKa = 10.3YY113 pKa = 10.69VEE115 pKa = 4.55QNVDD119 pKa = 3.23LQAQVAPNDD128 pKa = 3.24PRR130 pKa = 11.84YY131 pKa = 9.58RR132 pKa = 11.84DD133 pKa = 3.4QYY135 pKa = 11.93ADD137 pKa = 3.45QQVNAPEE144 pKa = 4.2AWDD147 pKa = 3.55ATFGDD152 pKa = 3.83SGVTVAVVDD161 pKa = 3.52QGVKK165 pKa = 10.09YY166 pKa = 10.07DD167 pKa = 4.85HH168 pKa = 7.72PDD170 pKa = 3.02LDD172 pKa = 4.3GNMDD176 pKa = 3.71GSVSNSGYY184 pKa = 11.13DD185 pKa = 4.3FVDD188 pKa = 5.06DD189 pKa = 6.14DD190 pKa = 5.45DD191 pKa = 6.29DD192 pKa = 5.04PYY194 pKa = 11.53PDD196 pKa = 4.25ALSDD200 pKa = 3.91EE201 pKa = 4.56YY202 pKa = 11.15HH203 pKa = 5.64GTHH206 pKa = 6.07VGGIAAAEE214 pKa = 4.02VDD216 pKa = 3.5NGEE219 pKa = 4.19GVTGIGNSSLLSGRR233 pKa = 11.84ALSEE237 pKa = 3.94QGSGSTADD245 pKa = 3.35IADD248 pKa = 3.88AVTWAADD255 pKa = 3.29QGADD259 pKa = 4.73VINLSLGGGGYY270 pKa = 8.89TDD272 pKa = 3.24TMKK275 pKa = 10.92NAVSYY280 pKa = 8.31ATNNGALVVAAAGNDD295 pKa = 3.82GQGSVSYY302 pKa = 9.27PAAYY306 pKa = 9.87SEE308 pKa = 4.45CLAVSALDD316 pKa = 3.77PDD318 pKa = 3.94EE319 pKa = 4.65TLASYY324 pKa = 11.31SNYY327 pKa = 10.19GGNIEE332 pKa = 4.42LAAPGTNVLSTWTDD346 pKa = 3.17DD347 pKa = 4.0AYY349 pKa = 11.12DD350 pKa = 4.39SISGTSMATPVVAGVAGLALAQWDD374 pKa = 4.01LTNAEE379 pKa = 4.73LRR381 pKa = 11.84NHH383 pKa = 7.19LKK385 pKa = 8.53NTAVNVGLSSDD396 pKa = 3.43KK397 pKa = 10.86QGSGRR402 pKa = 11.84VDD404 pKa = 3.09AGNAITTEE412 pKa = 4.31PGSGGGGGGGGGGSTSTTISGSLSSYY438 pKa = 10.99ADD440 pKa = 3.85SDD442 pKa = 3.81CSTYY446 pKa = 10.13TFEE449 pKa = 4.91YY450 pKa = 10.23DD451 pKa = 3.33SPSQIVIEE459 pKa = 4.16LSGPSDD465 pKa = 4.11ADD467 pKa = 3.05FDD469 pKa = 5.2LYY471 pKa = 11.65ANTGVGACPTTDD483 pKa = 3.35DD484 pKa = 3.72YY485 pKa = 11.81DD486 pKa = 3.87YY487 pKa = 11.49RR488 pKa = 11.84SYY490 pKa = 8.17TTNSQEE496 pKa = 4.43TITIDD501 pKa = 4.4SPDD504 pKa = 3.67TSTDD508 pKa = 2.99LYY510 pKa = 11.42ALVDD514 pKa = 4.1SYY516 pKa = 11.78SGSGDD521 pKa = 3.28FTVTFTEE528 pKa = 4.51YY529 pKa = 10.23QQ530 pKa = 3.16
Molecular weight: 54.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I0QUJ0|A0A1I0QUJ0_9EURY Acyl-CoA thioester hydrolase OS=Halobacterium jilantaiense OX=355548 GN=SAMN04487945_3013 PE=4 SV=1
MM1 pKa = 7.92DD2 pKa = 5.61PALAALFVGAALASLFMAWVIGAGSSGATPFAPAVGANAIATMRR46 pKa = 11.84AAFVVGLFGFAGAVTQGGSVSEE68 pKa = 4.52AIGGGLVGGASLPVAGVILVLLLGAGLMAVGIQTGYY104 pKa = 10.08PIATAFTVTGAVIGVGLALGGTPVWPKK131 pKa = 9.73YY132 pKa = 9.85RR133 pKa = 11.84QIAAVWALTPFVGGGIAFAIASVLPRR159 pKa = 11.84PGVPEE164 pKa = 4.55RR165 pKa = 11.84YY166 pKa = 9.2SVPVLAGLVAAVLANVEE183 pKa = 4.61FSFLGRR189 pKa = 11.84GASSGSVRR197 pKa = 11.84SLGRR201 pKa = 11.84QALPVGGPTAAAAVTGIAALAVGVVVWWDD230 pKa = 3.12VRR232 pKa = 11.84RR233 pKa = 11.84DD234 pKa = 3.39EE235 pKa = 4.81RR236 pKa = 11.84GGLRR240 pKa = 11.84RR241 pKa = 11.84VLLALGSLVAFSAGGSQVGLAVGPLLPLLDD271 pKa = 4.02EE272 pKa = 4.61VGGLSVFAVLVGGGLGMLVGSWTGAPRR299 pKa = 11.84MIKK302 pKa = 10.14SLAQDD307 pKa = 3.72YY308 pKa = 11.14SSLGPRR314 pKa = 11.84RR315 pKa = 11.84SISALVPSFLIAQLAVLLGVPVSFNEE341 pKa = 4.03IVVSAIIGSGAAVGGRR357 pKa = 11.84AAVDD361 pKa = 3.12ARR363 pKa = 11.84KK364 pKa = 9.76ILVTVGAWAGSFVLSFTLAYY384 pKa = 9.67VAAASLLL391 pKa = 3.87
MM1 pKa = 7.92DD2 pKa = 5.61PALAALFVGAALASLFMAWVIGAGSSGATPFAPAVGANAIATMRR46 pKa = 11.84AAFVVGLFGFAGAVTQGGSVSEE68 pKa = 4.52AIGGGLVGGASLPVAGVILVLLLGAGLMAVGIQTGYY104 pKa = 10.08PIATAFTVTGAVIGVGLALGGTPVWPKK131 pKa = 9.73YY132 pKa = 9.85RR133 pKa = 11.84QIAAVWALTPFVGGGIAFAIASVLPRR159 pKa = 11.84PGVPEE164 pKa = 4.55RR165 pKa = 11.84YY166 pKa = 9.2SVPVLAGLVAAVLANVEE183 pKa = 4.61FSFLGRR189 pKa = 11.84GASSGSVRR197 pKa = 11.84SLGRR201 pKa = 11.84QALPVGGPTAAAAVTGIAALAVGVVVWWDD230 pKa = 3.12VRR232 pKa = 11.84RR233 pKa = 11.84DD234 pKa = 3.39EE235 pKa = 4.81RR236 pKa = 11.84GGLRR240 pKa = 11.84RR241 pKa = 11.84VLLALGSLVAFSAGGSQVGLAVGPLLPLLDD271 pKa = 4.02EE272 pKa = 4.61VGGLSVFAVLVGGGLGMLVGSWTGAPRR299 pKa = 11.84MIKK302 pKa = 10.14SLAQDD307 pKa = 3.72YY308 pKa = 11.14SSLGPRR314 pKa = 11.84RR315 pKa = 11.84SISALVPSFLIAQLAVLLGVPVSFNEE341 pKa = 4.03IVVSAIIGSGAAVGGRR357 pKa = 11.84AAVDD361 pKa = 3.12ARR363 pKa = 11.84KK364 pKa = 9.76ILVTVGAWAGSFVLSFTLAYY384 pKa = 9.67VAAASLLL391 pKa = 3.87
Molecular weight: 38.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
884077 |
39 |
2195 |
289.5 |
31.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.059 ± 0.078 | 0.666 ± 0.014 |
9.186 ± 0.057 | 8.039 ± 0.069 |
3.277 ± 0.026 | 8.654 ± 0.046 |
2.028 ± 0.022 | 2.965 ± 0.037 |
1.628 ± 0.027 | 8.793 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.616 ± 0.018 | 2.181 ± 0.029 |
4.604 ± 0.032 | 2.491 ± 0.032 |
6.411 ± 0.044 | 5.405 ± 0.039 |
6.307 ± 0.045 | 9.944 ± 0.059 |
1.125 ± 0.016 | 2.621 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
