
Flavobacterium lacus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
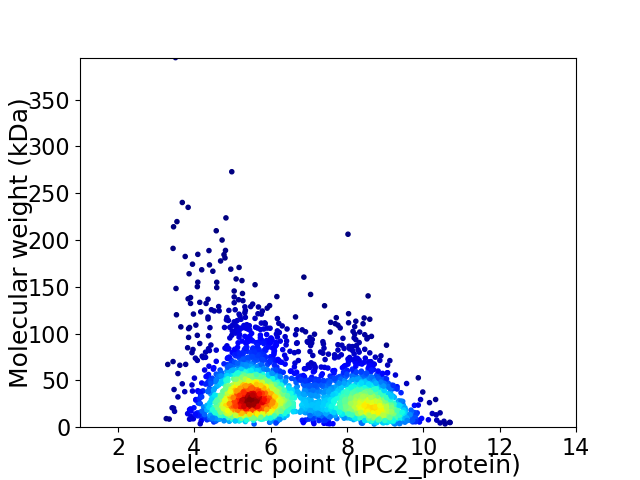
Virtual 2D-PAGE plot for 3053 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A328WSS7|A0A328WSS7_9FLAO Uncharacterized protein OS=Flavobacterium lacus OX=1353778 GN=B0I10_1083 PE=4 SV=1
MM1 pKa = 7.51KK2 pKa = 10.45KK3 pKa = 8.49ITNLFWLCLIFISSISFAQTARR25 pKa = 11.84VQVIHH30 pKa = 6.62NSPDD34 pKa = 3.23LAASSVDD41 pKa = 3.46VYY43 pKa = 11.66LNDD46 pKa = 3.62VLLLNDD52 pKa = 3.3FHH54 pKa = 7.28FRR56 pKa = 11.84TATPFIDD63 pKa = 3.86APAGVEE69 pKa = 3.83LSIDD73 pKa = 3.5IAPSSSSSSAEE84 pKa = 3.8SIYY87 pKa = 11.33NLTTTLSEE95 pKa = 4.09FEE97 pKa = 4.26TYY99 pKa = 10.24IIIANGIISPTGYY112 pKa = 10.31SPSTPFGLQVLVGGRR127 pKa = 11.84EE128 pKa = 4.2GTAVPDD134 pKa = 3.72TTDD137 pKa = 3.11VIVFHH142 pKa = 7.06GSTDD146 pKa = 3.32APTVDD151 pKa = 3.65VVEE154 pKa = 4.39LAVPAGTIVNDD165 pKa = 3.4ISYY168 pKa = 10.97GSFSEE173 pKa = 4.26YY174 pKa = 11.07LEE176 pKa = 5.33LPTNNYY182 pKa = 8.66VLAVRR187 pKa = 11.84DD188 pKa = 3.74EE189 pKa = 4.45TGTATVATYY198 pKa = 10.01SAPLADD204 pKa = 6.06LFLQTDD210 pKa = 5.22AITVLASGFLDD221 pKa = 4.23PSQNSNGPEE230 pKa = 3.57FGLWVALASGGDD242 pKa = 4.78LIPLPLFHH250 pKa = 7.08NPTARR255 pKa = 11.84LQVIHH260 pKa = 6.66NSPDD264 pKa = 3.25LAASAVDD271 pKa = 3.79VYY273 pKa = 12.02VNGALLLDD281 pKa = 3.65NFAFRR286 pKa = 11.84TATPFIDD293 pKa = 4.01VPAGIALSIDD303 pKa = 3.65VAPSTSTSSAEE314 pKa = 3.81SLYY317 pKa = 11.36NLTATLNEE325 pKa = 4.03FEE327 pKa = 4.7TYY329 pKa = 10.14IIVANGIVSPTGYY342 pKa = 10.38SPNQPFALEE351 pKa = 4.02VLVGGRR357 pKa = 11.84EE358 pKa = 4.14GTAVEE363 pKa = 4.48DD364 pKa = 3.86TTDD367 pKa = 3.35VIVFHH372 pKa = 6.82GSPDD376 pKa = 3.49APTVDD381 pKa = 3.49VVEE384 pKa = 4.46TSVPAGTLVDD394 pKa = 3.63NLDD397 pKa = 3.61YY398 pKa = 11.5GSFSNYY404 pKa = 10.25LEE406 pKa = 5.17LPTNNYY412 pKa = 8.97TIDD415 pKa = 3.43VRR417 pKa = 11.84DD418 pKa = 3.5EE419 pKa = 4.24TGTVTVATYY428 pKa = 9.97SAPLADD434 pKa = 6.22LFLQLDD440 pKa = 5.16AITVVASGFLDD451 pKa = 4.18PSQNSNGPAFGLWVAKK467 pKa = 9.25ATGGALIPLPLVEE480 pKa = 4.71TPTARR485 pKa = 11.84LQAIHH490 pKa = 6.57NSADD494 pKa = 3.2LAAALVDD501 pKa = 3.73VYY503 pKa = 11.9VNDD506 pKa = 3.92EE507 pKa = 4.09LLLNDD512 pKa = 4.06FAFRR516 pKa = 11.84TATPFIDD523 pKa = 3.66VPAGVEE529 pKa = 3.76LSIDD533 pKa = 3.76VAPSTSTSSAEE544 pKa = 4.04SIYY547 pKa = 11.3NLTATLAEE555 pKa = 4.45GEE557 pKa = 4.49TYY559 pKa = 10.54ILVANGIVSPTGYY572 pKa = 10.21SVAPNFAINVFAQGRR587 pKa = 11.84EE588 pKa = 3.99VAANSANTDD597 pKa = 3.19VLVNHH602 pKa = 7.34GSPDD606 pKa = 3.55APTVDD611 pKa = 3.49VVEE614 pKa = 4.29TSIPAGTIVDD624 pKa = 4.07NISFPEE630 pKa = 3.98FQGYY634 pKa = 10.53LDD636 pKa = 4.91LPNLDD641 pKa = 3.35FTLDD645 pKa = 3.5VRR647 pKa = 11.84DD648 pKa = 3.55EE649 pKa = 4.44TGTVTVASYY658 pKa = 9.19QAPLQTLGLEE668 pKa = 4.52GAAITVLASGFLDD681 pKa = 4.21PSANSNGPAFGLWVALAEE699 pKa = 4.63GGALIPLPLVEE710 pKa = 4.71TPTARR715 pKa = 11.84LQAIHH720 pKa = 6.59NSPDD724 pKa = 3.0LAAALVDD731 pKa = 3.73VYY733 pKa = 11.9VNDD736 pKa = 3.92EE737 pKa = 4.09LLLNDD742 pKa = 4.06FAFRR746 pKa = 11.84TATPFIDD753 pKa = 3.66VPAGVEE759 pKa = 3.76LSIDD763 pKa = 3.76VAPSTSTSSAEE774 pKa = 4.04SIYY777 pKa = 11.3NLTATLAEE785 pKa = 4.45GEE787 pKa = 4.49TYY789 pKa = 10.54ILVANGIVSPTGYY802 pKa = 10.27SVGPNFAINVFAQGRR817 pKa = 11.84EE818 pKa = 3.99VAANSANTDD827 pKa = 3.19VLVNHH832 pKa = 7.34GSPDD836 pKa = 3.55APTVDD841 pKa = 3.49VVEE844 pKa = 4.29TSIPAGTIVNNISFPEE860 pKa = 4.0FQGYY864 pKa = 10.53LDD866 pKa = 4.91LPNLDD871 pKa = 3.35FTLDD875 pKa = 3.5VRR877 pKa = 11.84DD878 pKa = 3.55EE879 pKa = 4.44TGTVTVASYY888 pKa = 9.19QAPLQTLGLEE898 pKa = 4.52GAAITVLASGFLDD911 pKa = 4.21PSANSNGPAFGLWVALASGGDD932 pKa = 4.63LIPLPEE938 pKa = 5.26IPLNVDD944 pKa = 3.3SFNLNSISLFPNPAKK959 pKa = 9.79EE960 pKa = 4.34TINLSIPFDD969 pKa = 3.85YY970 pKa = 11.1EE971 pKa = 3.87NFSATIYY978 pKa = 10.4DD979 pKa = 3.68LSGRR983 pKa = 11.84LVKK986 pKa = 9.83TVTTPTDD993 pKa = 3.07IDD995 pKa = 3.35ISEE998 pKa = 4.69LNSGLYY1004 pKa = 9.79ILNTTLNDD1012 pKa = 3.38LSFNQKK1018 pKa = 8.42FVKK1021 pKa = 10.35NN1022 pKa = 3.81
MM1 pKa = 7.51KK2 pKa = 10.45KK3 pKa = 8.49ITNLFWLCLIFISSISFAQTARR25 pKa = 11.84VQVIHH30 pKa = 6.62NSPDD34 pKa = 3.23LAASSVDD41 pKa = 3.46VYY43 pKa = 11.66LNDD46 pKa = 3.62VLLLNDD52 pKa = 3.3FHH54 pKa = 7.28FRR56 pKa = 11.84TATPFIDD63 pKa = 3.86APAGVEE69 pKa = 3.83LSIDD73 pKa = 3.5IAPSSSSSSAEE84 pKa = 3.8SIYY87 pKa = 11.33NLTTTLSEE95 pKa = 4.09FEE97 pKa = 4.26TYY99 pKa = 10.24IIIANGIISPTGYY112 pKa = 10.31SPSTPFGLQVLVGGRR127 pKa = 11.84EE128 pKa = 4.2GTAVPDD134 pKa = 3.72TTDD137 pKa = 3.11VIVFHH142 pKa = 7.06GSTDD146 pKa = 3.32APTVDD151 pKa = 3.65VVEE154 pKa = 4.39LAVPAGTIVNDD165 pKa = 3.4ISYY168 pKa = 10.97GSFSEE173 pKa = 4.26YY174 pKa = 11.07LEE176 pKa = 5.33LPTNNYY182 pKa = 8.66VLAVRR187 pKa = 11.84DD188 pKa = 3.74EE189 pKa = 4.45TGTATVATYY198 pKa = 10.01SAPLADD204 pKa = 6.06LFLQTDD210 pKa = 5.22AITVLASGFLDD221 pKa = 4.23PSQNSNGPEE230 pKa = 3.57FGLWVALASGGDD242 pKa = 4.78LIPLPLFHH250 pKa = 7.08NPTARR255 pKa = 11.84LQVIHH260 pKa = 6.66NSPDD264 pKa = 3.25LAASAVDD271 pKa = 3.79VYY273 pKa = 12.02VNGALLLDD281 pKa = 3.65NFAFRR286 pKa = 11.84TATPFIDD293 pKa = 4.01VPAGIALSIDD303 pKa = 3.65VAPSTSTSSAEE314 pKa = 3.81SLYY317 pKa = 11.36NLTATLNEE325 pKa = 4.03FEE327 pKa = 4.7TYY329 pKa = 10.14IIVANGIVSPTGYY342 pKa = 10.38SPNQPFALEE351 pKa = 4.02VLVGGRR357 pKa = 11.84EE358 pKa = 4.14GTAVEE363 pKa = 4.48DD364 pKa = 3.86TTDD367 pKa = 3.35VIVFHH372 pKa = 6.82GSPDD376 pKa = 3.49APTVDD381 pKa = 3.49VVEE384 pKa = 4.46TSVPAGTLVDD394 pKa = 3.63NLDD397 pKa = 3.61YY398 pKa = 11.5GSFSNYY404 pKa = 10.25LEE406 pKa = 5.17LPTNNYY412 pKa = 8.97TIDD415 pKa = 3.43VRR417 pKa = 11.84DD418 pKa = 3.5EE419 pKa = 4.24TGTVTVATYY428 pKa = 9.97SAPLADD434 pKa = 6.22LFLQLDD440 pKa = 5.16AITVVASGFLDD451 pKa = 4.18PSQNSNGPAFGLWVAKK467 pKa = 9.25ATGGALIPLPLVEE480 pKa = 4.71TPTARR485 pKa = 11.84LQAIHH490 pKa = 6.57NSADD494 pKa = 3.2LAAALVDD501 pKa = 3.73VYY503 pKa = 11.9VNDD506 pKa = 3.92EE507 pKa = 4.09LLLNDD512 pKa = 4.06FAFRR516 pKa = 11.84TATPFIDD523 pKa = 3.66VPAGVEE529 pKa = 3.76LSIDD533 pKa = 3.76VAPSTSTSSAEE544 pKa = 4.04SIYY547 pKa = 11.3NLTATLAEE555 pKa = 4.45GEE557 pKa = 4.49TYY559 pKa = 10.54ILVANGIVSPTGYY572 pKa = 10.21SVAPNFAINVFAQGRR587 pKa = 11.84EE588 pKa = 3.99VAANSANTDD597 pKa = 3.19VLVNHH602 pKa = 7.34GSPDD606 pKa = 3.55APTVDD611 pKa = 3.49VVEE614 pKa = 4.29TSIPAGTIVDD624 pKa = 4.07NISFPEE630 pKa = 3.98FQGYY634 pKa = 10.53LDD636 pKa = 4.91LPNLDD641 pKa = 3.35FTLDD645 pKa = 3.5VRR647 pKa = 11.84DD648 pKa = 3.55EE649 pKa = 4.44TGTVTVASYY658 pKa = 9.19QAPLQTLGLEE668 pKa = 4.52GAAITVLASGFLDD681 pKa = 4.21PSANSNGPAFGLWVALAEE699 pKa = 4.63GGALIPLPLVEE710 pKa = 4.71TPTARR715 pKa = 11.84LQAIHH720 pKa = 6.59NSPDD724 pKa = 3.0LAAALVDD731 pKa = 3.73VYY733 pKa = 11.9VNDD736 pKa = 3.92EE737 pKa = 4.09LLLNDD742 pKa = 4.06FAFRR746 pKa = 11.84TATPFIDD753 pKa = 3.66VPAGVEE759 pKa = 3.76LSIDD763 pKa = 3.76VAPSTSTSSAEE774 pKa = 4.04SIYY777 pKa = 11.3NLTATLAEE785 pKa = 4.45GEE787 pKa = 4.49TYY789 pKa = 10.54ILVANGIVSPTGYY802 pKa = 10.27SVGPNFAINVFAQGRR817 pKa = 11.84EE818 pKa = 3.99VAANSANTDD827 pKa = 3.19VLVNHH832 pKa = 7.34GSPDD836 pKa = 3.55APTVDD841 pKa = 3.49VVEE844 pKa = 4.29TSIPAGTIVNNISFPEE860 pKa = 4.0FQGYY864 pKa = 10.53LDD866 pKa = 4.91LPNLDD871 pKa = 3.35FTLDD875 pKa = 3.5VRR877 pKa = 11.84DD878 pKa = 3.55EE879 pKa = 4.44TGTVTVASYY888 pKa = 9.19QAPLQTLGLEE898 pKa = 4.52GAAITVLASGFLDD911 pKa = 4.21PSANSNGPAFGLWVALASGGDD932 pKa = 4.63LIPLPEE938 pKa = 5.26IPLNVDD944 pKa = 3.3SFNLNSISLFPNPAKK959 pKa = 9.79EE960 pKa = 4.34TINLSIPFDD969 pKa = 3.85YY970 pKa = 11.1EE971 pKa = 3.87NFSATIYY978 pKa = 10.4DD979 pKa = 3.68LSGRR983 pKa = 11.84LVKK986 pKa = 9.83TVTTPTDD993 pKa = 3.07IDD995 pKa = 3.35ISEE998 pKa = 4.69LNSGLYY1004 pKa = 9.79ILNTTLNDD1012 pKa = 3.38LSFNQKK1018 pKa = 8.42FVKK1021 pKa = 10.35NN1022 pKa = 3.81
Molecular weight: 107.2 kDa
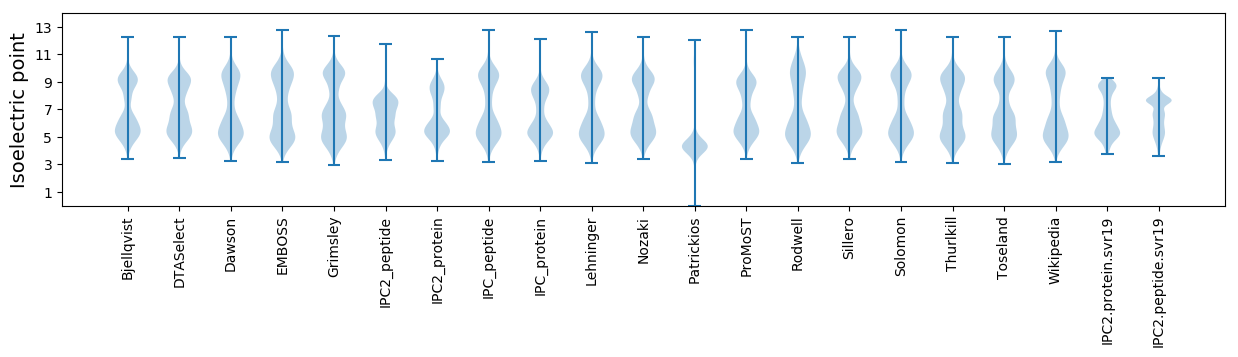
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A328WLQ6|A0A328WLQ6_9FLAO N-dimethylarginine dimethylaminohydrolase OS=Flavobacterium lacus OX=1353778 GN=B0I10_111100 PE=4 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.39KK31 pKa = 10.56SLLAPKK37 pKa = 10.25NNSGGRR43 pKa = 11.84NSQGKK48 pKa = 6.33MTMRR52 pKa = 11.84YY53 pKa = 7.39TGGGHH58 pKa = 4.44KK59 pKa = 9.52QKK61 pKa = 10.78YY62 pKa = 9.58RR63 pKa = 11.84IIDD66 pKa = 3.78FKK68 pKa = 10.28RR69 pKa = 11.84TKK71 pKa = 10.51DD72 pKa = 3.79GIPATVKK79 pKa = 10.64SIEE82 pKa = 4.0YY83 pKa = 10.15DD84 pKa = 3.43PNRR87 pKa = 11.84TAFIALLAYY96 pKa = 10.34ADD98 pKa = 3.82GAKK101 pKa = 9.85TYY103 pKa = 10.97VIAQNGLNVGQTVTSGEE120 pKa = 4.06GSAPEE125 pKa = 4.24IGNALPLSKK134 pKa = 10.33IPLGTVISCIEE145 pKa = 3.9LRR147 pKa = 11.84PGQGAIIARR156 pKa = 11.84SAGTFAQLMARR167 pKa = 11.84DD168 pKa = 4.06GKK170 pKa = 10.75YY171 pKa = 9.05ATIKK175 pKa = 9.6MPSGEE180 pKa = 4.18TRR182 pKa = 11.84LILLTCMATIGAVSNSDD199 pKa = 2.96HH200 pKa = 6.02QLIVSGKK207 pKa = 8.58AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVAMNPVDD229 pKa = 3.63HH230 pKa = 7.13PMGGGEE236 pKa = 4.08GRR238 pKa = 11.84SSGGHH243 pKa = 4.7PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.29GYY255 pKa = 6.86RR256 pKa = 11.84TRR258 pKa = 11.84SKK260 pKa = 10.63VSPSNKK266 pKa = 9.58YY267 pKa = 9.24IVEE270 pKa = 4.07RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.24KK274 pKa = 10.1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGFDD22 pKa = 5.49AITTDD27 pKa = 3.56KK28 pKa = 11.07PEE30 pKa = 4.39KK31 pKa = 10.56SLLAPKK37 pKa = 10.25NNSGGRR43 pKa = 11.84NSQGKK48 pKa = 6.33MTMRR52 pKa = 11.84YY53 pKa = 7.39TGGGHH58 pKa = 4.44KK59 pKa = 9.52QKK61 pKa = 10.78YY62 pKa = 9.58RR63 pKa = 11.84IIDD66 pKa = 3.78FKK68 pKa = 10.28RR69 pKa = 11.84TKK71 pKa = 10.51DD72 pKa = 3.79GIPATVKK79 pKa = 10.64SIEE82 pKa = 4.0YY83 pKa = 10.15DD84 pKa = 3.43PNRR87 pKa = 11.84TAFIALLAYY96 pKa = 10.34ADD98 pKa = 3.82GAKK101 pKa = 9.85TYY103 pKa = 10.97VIAQNGLNVGQTVTSGEE120 pKa = 4.06GSAPEE125 pKa = 4.24IGNALPLSKK134 pKa = 10.33IPLGTVISCIEE145 pKa = 3.9LRR147 pKa = 11.84PGQGAIIARR156 pKa = 11.84SAGTFAQLMARR167 pKa = 11.84DD168 pKa = 4.06GKK170 pKa = 10.75YY171 pKa = 9.05ATIKK175 pKa = 9.6MPSGEE180 pKa = 4.18TRR182 pKa = 11.84LILLTCMATIGAVSNSDD199 pKa = 2.96HH200 pKa = 6.02QLIVSGKK207 pKa = 8.58AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVAMNPVDD229 pKa = 3.63HH230 pKa = 7.13PMGGGEE236 pKa = 4.08GRR238 pKa = 11.84SSGGHH243 pKa = 4.7PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.29GYY255 pKa = 6.86RR256 pKa = 11.84TRR258 pKa = 11.84SKK260 pKa = 10.63VSPSNKK266 pKa = 9.58YY267 pKa = 9.24IVEE270 pKa = 4.07RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.24KK274 pKa = 10.1
Molecular weight: 29.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1057804 |
29 |
3746 |
346.5 |
39.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.175 ± 0.047 | 0.791 ± 0.017 |
5.147 ± 0.033 | 6.512 ± 0.059 |
5.648 ± 0.039 | 6.28 ± 0.06 |
1.662 ± 0.022 | 8.081 ± 0.044 |
7.544 ± 0.085 | 9.153 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.144 ± 0.024 | 6.594 ± 0.05 |
3.439 ± 0.035 | 3.578 ± 0.024 |
3.148 ± 0.032 | 6.599 ± 0.044 |
6.173 ± 0.087 | 6.377 ± 0.038 |
1.012 ± 0.017 | 3.945 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
