
Camelus dromedarius papillomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Deltapapillomavirus; Deltapapillomavirus 6
Average proteome isoelectric point is 7.5
Get precalculated fractions of proteins
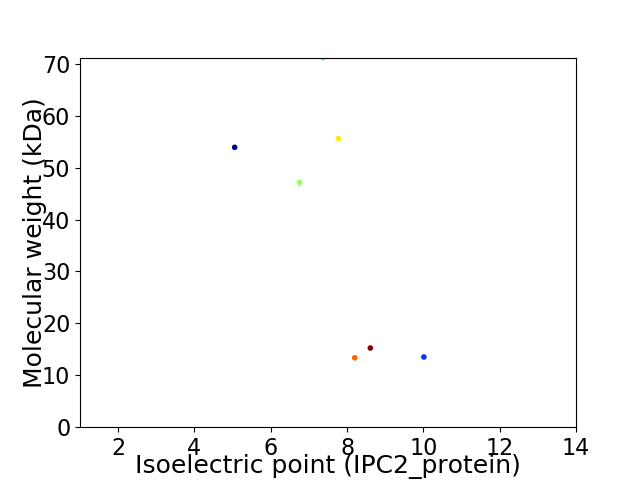
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F2YGI1|F2YGI1_9PAPI Major capsid protein L1 OS=Camelus dromedarius papillomavirus 2 OX=996651 GN=L1 PE=3 SV=1
MM1 pKa = 7.83ASRR4 pKa = 11.84KK5 pKa = 8.71RR6 pKa = 11.84VRR8 pKa = 11.84RR9 pKa = 11.84ANPYY13 pKa = 9.77EE14 pKa = 3.99LYY16 pKa = 8.96RR17 pKa = 11.84TCKK20 pKa = 9.96AANTCPRR27 pKa = 11.84DD28 pKa = 3.74VIPLVEE34 pKa = 4.69GNTLADD40 pKa = 4.74QILKK44 pKa = 8.1WGSWAVYY51 pKa = 10.18LGGLGIGTGTQSISGRR67 pKa = 11.84PITTGSVKK75 pKa = 10.89SNIEE79 pKa = 3.81KK80 pKa = 10.63FLVNLFGKK88 pKa = 10.7GGTQTRR94 pKa = 11.84APTGGIPLEE103 pKa = 4.12TLGAGTSLGTPDD115 pKa = 3.55VAVTATDD122 pKa = 3.33APTVVVPEE130 pKa = 4.4APSGGEE136 pKa = 3.73PGEE139 pKa = 4.07GLGVVTEE146 pKa = 4.38NTQEE150 pKa = 4.0TAFTYY155 pKa = 10.93VDD157 pKa = 3.25VDD159 pKa = 3.66DD160 pKa = 4.5TGEE163 pKa = 3.91IMVVNLRR170 pKa = 11.84PDD172 pKa = 3.69GDD174 pKa = 4.61DD175 pKa = 3.47PTSHH179 pKa = 7.1LSSSTTFHH187 pKa = 6.5NPTYY191 pKa = 9.97QEE193 pKa = 3.56PSGRR197 pKa = 11.84LEE199 pKa = 4.42TVGEE203 pKa = 4.18TSNTEE208 pKa = 4.16NIFLAGEE215 pKa = 4.22NLGSTGGEE223 pKa = 3.96EE224 pKa = 4.05IEE226 pKa = 4.86LSLFSKK232 pKa = 10.42PRR234 pKa = 11.84TSTPRR239 pKa = 11.84QSPRR243 pKa = 11.84APVRR247 pKa = 11.84GRR249 pKa = 11.84ANWFSRR255 pKa = 11.84RR256 pKa = 11.84YY257 pKa = 7.69YY258 pKa = 9.53TQLNLNNPAVITQPGEE274 pKa = 3.96VIGTQFVNPVFEE286 pKa = 4.64GSSFEE291 pKa = 4.4APPAYY296 pKa = 9.84EE297 pKa = 3.9SWNTEE302 pKa = 3.82LPEE305 pKa = 4.67LRR307 pKa = 11.84DD308 pKa = 3.63ATHH311 pKa = 5.48YY312 pKa = 7.7TASRR316 pKa = 11.84LLQGPSGRR324 pKa = 11.84VGVDD328 pKa = 3.14RR329 pKa = 11.84VATTRR334 pKa = 11.84TIGTRR339 pKa = 11.84SGIRR343 pKa = 11.84IGPEE347 pKa = 3.3VHH349 pKa = 6.1VRR351 pKa = 11.84QSLSSISDD359 pKa = 3.14AVEE362 pKa = 3.6FATFVSGDD370 pKa = 3.54VEE372 pKa = 4.22TAPTVEE378 pKa = 4.07TAFVEE383 pKa = 4.66EE384 pKa = 4.27TAPFEE389 pKa = 6.17DD390 pKa = 3.64IDD392 pKa = 4.49LDD394 pKa = 4.15SVYY397 pKa = 11.03SDD399 pKa = 4.39EE400 pKa = 5.95DD401 pKa = 4.19LLDD404 pKa = 3.61QYY406 pKa = 11.23PATSHH411 pKa = 5.42GRR413 pKa = 11.84LVFGEE418 pKa = 4.18RR419 pKa = 11.84FNRR422 pKa = 11.84DD423 pKa = 2.88IVPIPIDD430 pKa = 3.71PPFTALVPVDD440 pKa = 3.24VTTYY444 pKa = 7.86VTPGANWPDD453 pKa = 3.64FTPGSKK459 pKa = 10.49GGMTPGIIIDD469 pKa = 3.86LSEE472 pKa = 4.22EE473 pKa = 4.21LFKK476 pKa = 10.68HH477 pKa = 5.86YY478 pKa = 10.29YY479 pKa = 9.25LHH481 pKa = 7.39EE482 pKa = 4.43SLLRR486 pKa = 11.84RR487 pKa = 11.84KK488 pKa = 9.46RR489 pKa = 11.84RR490 pKa = 11.84RR491 pKa = 11.84RR492 pKa = 11.84KK493 pKa = 9.54RR494 pKa = 11.84LYY496 pKa = 10.37AA497 pKa = 4.23
MM1 pKa = 7.83ASRR4 pKa = 11.84KK5 pKa = 8.71RR6 pKa = 11.84VRR8 pKa = 11.84RR9 pKa = 11.84ANPYY13 pKa = 9.77EE14 pKa = 3.99LYY16 pKa = 8.96RR17 pKa = 11.84TCKK20 pKa = 9.96AANTCPRR27 pKa = 11.84DD28 pKa = 3.74VIPLVEE34 pKa = 4.69GNTLADD40 pKa = 4.74QILKK44 pKa = 8.1WGSWAVYY51 pKa = 10.18LGGLGIGTGTQSISGRR67 pKa = 11.84PITTGSVKK75 pKa = 10.89SNIEE79 pKa = 3.81KK80 pKa = 10.63FLVNLFGKK88 pKa = 10.7GGTQTRR94 pKa = 11.84APTGGIPLEE103 pKa = 4.12TLGAGTSLGTPDD115 pKa = 3.55VAVTATDD122 pKa = 3.33APTVVVPEE130 pKa = 4.4APSGGEE136 pKa = 3.73PGEE139 pKa = 4.07GLGVVTEE146 pKa = 4.38NTQEE150 pKa = 4.0TAFTYY155 pKa = 10.93VDD157 pKa = 3.25VDD159 pKa = 3.66DD160 pKa = 4.5TGEE163 pKa = 3.91IMVVNLRR170 pKa = 11.84PDD172 pKa = 3.69GDD174 pKa = 4.61DD175 pKa = 3.47PTSHH179 pKa = 7.1LSSSTTFHH187 pKa = 6.5NPTYY191 pKa = 9.97QEE193 pKa = 3.56PSGRR197 pKa = 11.84LEE199 pKa = 4.42TVGEE203 pKa = 4.18TSNTEE208 pKa = 4.16NIFLAGEE215 pKa = 4.22NLGSTGGEE223 pKa = 3.96EE224 pKa = 4.05IEE226 pKa = 4.86LSLFSKK232 pKa = 10.42PRR234 pKa = 11.84TSTPRR239 pKa = 11.84QSPRR243 pKa = 11.84APVRR247 pKa = 11.84GRR249 pKa = 11.84ANWFSRR255 pKa = 11.84RR256 pKa = 11.84YY257 pKa = 7.69YY258 pKa = 9.53TQLNLNNPAVITQPGEE274 pKa = 3.96VIGTQFVNPVFEE286 pKa = 4.64GSSFEE291 pKa = 4.4APPAYY296 pKa = 9.84EE297 pKa = 3.9SWNTEE302 pKa = 3.82LPEE305 pKa = 4.67LRR307 pKa = 11.84DD308 pKa = 3.63ATHH311 pKa = 5.48YY312 pKa = 7.7TASRR316 pKa = 11.84LLQGPSGRR324 pKa = 11.84VGVDD328 pKa = 3.14RR329 pKa = 11.84VATTRR334 pKa = 11.84TIGTRR339 pKa = 11.84SGIRR343 pKa = 11.84IGPEE347 pKa = 3.3VHH349 pKa = 6.1VRR351 pKa = 11.84QSLSSISDD359 pKa = 3.14AVEE362 pKa = 3.6FATFVSGDD370 pKa = 3.54VEE372 pKa = 4.22TAPTVEE378 pKa = 4.07TAFVEE383 pKa = 4.66EE384 pKa = 4.27TAPFEE389 pKa = 6.17DD390 pKa = 3.64IDD392 pKa = 4.49LDD394 pKa = 4.15SVYY397 pKa = 11.03SDD399 pKa = 4.39EE400 pKa = 5.95DD401 pKa = 4.19LLDD404 pKa = 3.61QYY406 pKa = 11.23PATSHH411 pKa = 5.42GRR413 pKa = 11.84LVFGEE418 pKa = 4.18RR419 pKa = 11.84FNRR422 pKa = 11.84DD423 pKa = 2.88IVPIPIDD430 pKa = 3.71PPFTALVPVDD440 pKa = 3.24VTTYY444 pKa = 7.86VTPGANWPDD453 pKa = 3.64FTPGSKK459 pKa = 10.49GGMTPGIIIDD469 pKa = 3.86LSEE472 pKa = 4.22EE473 pKa = 4.21LFKK476 pKa = 10.68HH477 pKa = 5.86YY478 pKa = 10.29YY479 pKa = 9.25LHH481 pKa = 7.39EE482 pKa = 4.43SLLRR486 pKa = 11.84RR487 pKa = 11.84KK488 pKa = 9.46RR489 pKa = 11.84RR490 pKa = 11.84RR491 pKa = 11.84RR492 pKa = 11.84KK493 pKa = 9.54RR494 pKa = 11.84LYY496 pKa = 10.37AA497 pKa = 4.23
Molecular weight: 53.93 kDa
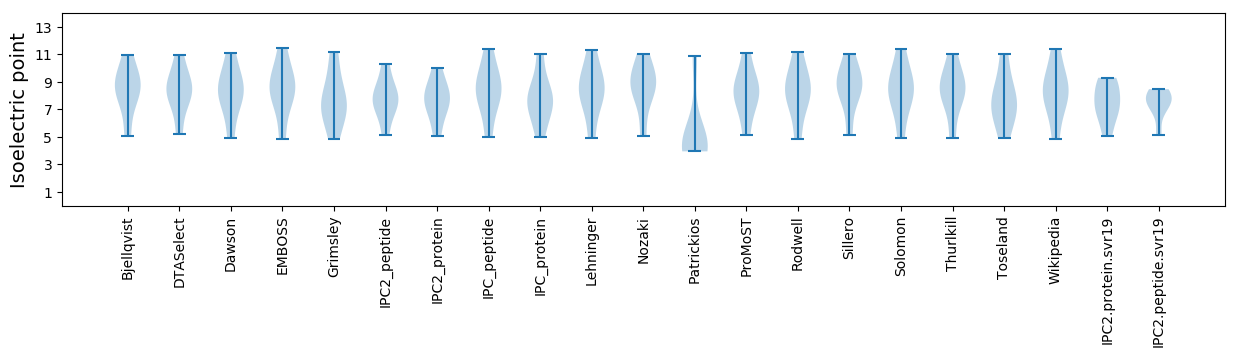
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2YGH8|F2YGH8_9PAPI Replication protein E1 OS=Camelus dromedarius papillomavirus 2 OX=996651 GN=E1 PE=3 SV=1
MM1 pKa = 7.49AAAEE5 pKa = 4.3DD6 pKa = 4.22TPPRR10 pKa = 11.84PGPSSRR16 pKa = 11.84CPPSIAPTVKK26 pKa = 9.97HH27 pKa = 6.13RR28 pKa = 11.84CPACKK33 pKa = 9.68GVHH36 pKa = 6.07GPITYY41 pKa = 8.44PQLPCLSLIKK51 pKa = 9.83WNPPTWVPSEE61 pKa = 4.26SAVLGPLHH69 pKa = 6.1VLRR72 pKa = 11.84PSRR75 pKa = 11.84YY76 pKa = 8.46YY77 pKa = 10.61GAQGSRR83 pKa = 11.84NAYY86 pKa = 9.8LPPPTCRR93 pKa = 11.84ARR95 pKa = 11.84SRR97 pKa = 11.84KK98 pKa = 8.56VWHH101 pKa = 6.13EE102 pKa = 3.25HH103 pKa = 5.23RR104 pKa = 11.84KK105 pKa = 7.29WMKK108 pKa = 9.69RR109 pKa = 11.84RR110 pKa = 11.84HH111 pKa = 5.5LRR113 pKa = 11.84TPRR116 pKa = 11.84RR117 pKa = 11.84YY118 pKa = 8.97PP119 pKa = 3.2
MM1 pKa = 7.49AAAEE5 pKa = 4.3DD6 pKa = 4.22TPPRR10 pKa = 11.84PGPSSRR16 pKa = 11.84CPPSIAPTVKK26 pKa = 9.97HH27 pKa = 6.13RR28 pKa = 11.84CPACKK33 pKa = 9.68GVHH36 pKa = 6.07GPITYY41 pKa = 8.44PQLPCLSLIKK51 pKa = 9.83WNPPTWVPSEE61 pKa = 4.26SAVLGPLHH69 pKa = 6.1VLRR72 pKa = 11.84PSRR75 pKa = 11.84YY76 pKa = 8.46YY77 pKa = 10.61GAQGSRR83 pKa = 11.84NAYY86 pKa = 9.8LPPPTCRR93 pKa = 11.84ARR95 pKa = 11.84SRR97 pKa = 11.84KK98 pKa = 8.56VWHH101 pKa = 6.13EE102 pKa = 3.25HH103 pKa = 5.23RR104 pKa = 11.84KK105 pKa = 7.29WMKK108 pKa = 9.69RR109 pKa = 11.84RR110 pKa = 11.84HH111 pKa = 5.5LRR113 pKa = 11.84TPRR116 pKa = 11.84RR117 pKa = 11.84YY118 pKa = 8.97PP119 pKa = 3.2
Molecular weight: 13.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2432 |
119 |
642 |
347.4 |
38.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.401 ± 0.887 | 2.426 ± 0.588 |
5.222 ± 0.32 | 5.387 ± 0.472 |
3.454 ± 0.408 | 7.196 ± 0.702 |
2.22 ± 0.32 | 3.331 ± 0.382 |
5.428 ± 0.828 | 7.936 ± 0.326 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.521 ± 0.248 | 3.824 ± 0.492 |
7.319 ± 1.334 | 4.235 ± 0.539 |
6.414 ± 0.717 | 7.689 ± 0.716 |
6.867 ± 1.013 | 6.826 ± 0.406 |
1.521 ± 0.218 | 3.783 ± 0.28 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
