
Nora virus
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
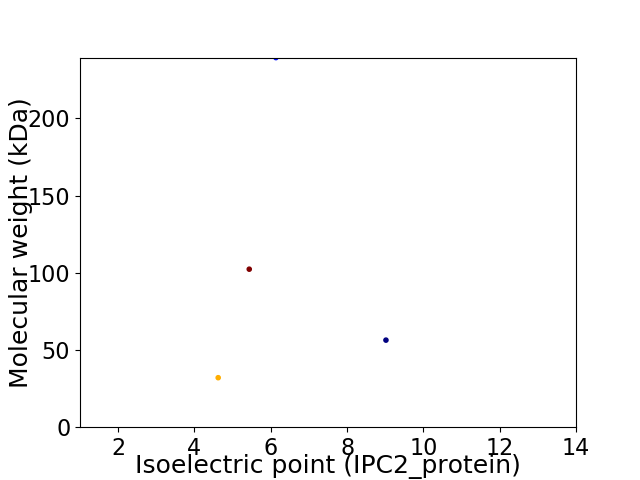
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D2WFA0|D2WFA0_NORAV Capsid protein OS=Nora virus OX=363716 PE=1 SV=1
MM1 pKa = 7.49ALKK4 pKa = 10.69EE5 pKa = 4.29EE6 pKa = 4.59IFDD9 pKa = 4.12QNTTLFAVLDD19 pKa = 4.21EE20 pKa = 4.76NEE22 pKa = 3.99VTEE25 pKa = 4.41IKK27 pKa = 10.44SIQASVTAVKK37 pKa = 8.71TQLDD41 pKa = 3.88QQKK44 pKa = 10.19LQLDD48 pKa = 4.31GLAKK52 pKa = 10.68VVDD55 pKa = 3.81NNQARR60 pKa = 11.84NEE62 pKa = 4.02EE63 pKa = 4.09QFVNINTTLVEE74 pKa = 4.06MSLEE78 pKa = 4.05VDD80 pKa = 3.28KK81 pKa = 10.86LTTTTSQQAKK91 pKa = 9.66QINTFATALNEE102 pKa = 3.88LDD104 pKa = 3.22QTTRR108 pKa = 11.84DD109 pKa = 3.71SLDD112 pKa = 3.41TLNTTTEE119 pKa = 4.2SLTKK123 pKa = 10.19QVLFNTDD130 pKa = 3.53EE131 pKa = 4.11ITVLKK136 pKa = 10.7VDD138 pKa = 3.81VATVTQKK145 pKa = 10.26QQDD148 pKa = 3.96VEE150 pKa = 4.42HH151 pKa = 7.19SLEE154 pKa = 4.15TMKK157 pKa = 11.23DD158 pKa = 3.57EE159 pKa = 4.45IGEE162 pKa = 4.05LHH164 pKa = 6.48VSVNANVNSIEE175 pKa = 3.97ALRR178 pKa = 11.84TRR180 pKa = 11.84IAALEE185 pKa = 3.98IRR187 pKa = 11.84DD188 pKa = 3.72VGPWVLKK195 pKa = 10.26DD196 pKa = 4.12RR197 pKa = 11.84IYY199 pKa = 11.25KK200 pKa = 9.59FVINKK205 pKa = 9.37PNGTTRR211 pKa = 11.84YY212 pKa = 3.95TTIYY216 pKa = 10.02FFADD220 pKa = 3.57VYY222 pKa = 11.19YY223 pKa = 9.58STGVRR228 pKa = 11.84AAPTNSGTATSILTITSLTTSYY250 pKa = 11.52SLANVPVLKK259 pKa = 10.42GVPYY263 pKa = 10.4RR264 pKa = 11.84VNGYY268 pKa = 8.54FANGNNIEE276 pKa = 5.34DD277 pKa = 3.84ITGSTSVIYY286 pKa = 10.68DD287 pKa = 3.4SMM289 pKa = 5.66
MM1 pKa = 7.49ALKK4 pKa = 10.69EE5 pKa = 4.29EE6 pKa = 4.59IFDD9 pKa = 4.12QNTTLFAVLDD19 pKa = 4.21EE20 pKa = 4.76NEE22 pKa = 3.99VTEE25 pKa = 4.41IKK27 pKa = 10.44SIQASVTAVKK37 pKa = 8.71TQLDD41 pKa = 3.88QQKK44 pKa = 10.19LQLDD48 pKa = 4.31GLAKK52 pKa = 10.68VVDD55 pKa = 3.81NNQARR60 pKa = 11.84NEE62 pKa = 4.02EE63 pKa = 4.09QFVNINTTLVEE74 pKa = 4.06MSLEE78 pKa = 4.05VDD80 pKa = 3.28KK81 pKa = 10.86LTTTTSQQAKK91 pKa = 9.66QINTFATALNEE102 pKa = 3.88LDD104 pKa = 3.22QTTRR108 pKa = 11.84DD109 pKa = 3.71SLDD112 pKa = 3.41TLNTTTEE119 pKa = 4.2SLTKK123 pKa = 10.19QVLFNTDD130 pKa = 3.53EE131 pKa = 4.11ITVLKK136 pKa = 10.7VDD138 pKa = 3.81VATVTQKK145 pKa = 10.26QQDD148 pKa = 3.96VEE150 pKa = 4.42HH151 pKa = 7.19SLEE154 pKa = 4.15TMKK157 pKa = 11.23DD158 pKa = 3.57EE159 pKa = 4.45IGEE162 pKa = 4.05LHH164 pKa = 6.48VSVNANVNSIEE175 pKa = 3.97ALRR178 pKa = 11.84TRR180 pKa = 11.84IAALEE185 pKa = 3.98IRR187 pKa = 11.84DD188 pKa = 3.72VGPWVLKK195 pKa = 10.26DD196 pKa = 4.12RR197 pKa = 11.84IYY199 pKa = 11.25KK200 pKa = 9.59FVINKK205 pKa = 9.37PNGTTRR211 pKa = 11.84YY212 pKa = 3.95TTIYY216 pKa = 10.02FFADD220 pKa = 3.57VYY222 pKa = 11.19YY223 pKa = 9.58STGVRR228 pKa = 11.84AAPTNSGTATSILTITSLTTSYY250 pKa = 11.52SLANVPVLKK259 pKa = 10.42GVPYY263 pKa = 10.4RR264 pKa = 11.84VNGYY268 pKa = 8.54FANGNNIEE276 pKa = 5.34DD277 pKa = 3.84ITGSTSVIYY286 pKa = 10.68DD287 pKa = 3.4SMM289 pKa = 5.66
Molecular weight: 32.05 kDa
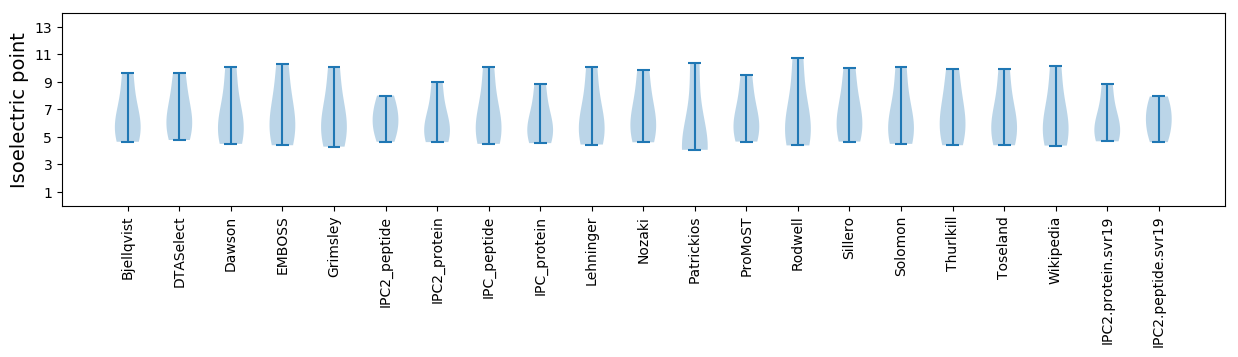
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D2WF98|D2WF98_NORAV Replication polyprotein OS=Nora virus OX=363716 PE=4 SV=1
MM1 pKa = 7.82INNQTNKK8 pKa = 10.17KK9 pKa = 9.21GPQLEE14 pKa = 4.1RR15 pKa = 11.84VHH17 pKa = 6.82FGSTQVVGKK26 pKa = 7.26STKK29 pKa = 9.43RR30 pKa = 11.84RR31 pKa = 11.84QRR33 pKa = 11.84GTKK36 pKa = 9.87LDD38 pKa = 2.94IEE40 pKa = 4.57YY41 pKa = 9.64TVRR44 pKa = 11.84RR45 pKa = 11.84NDD47 pKa = 3.13APKK50 pKa = 9.57EE51 pKa = 3.95QKK53 pKa = 10.53FLISEE58 pKa = 4.1IFDD61 pKa = 3.51EE62 pKa = 5.62KK63 pKa = 10.92LDD65 pKa = 3.78KK66 pKa = 10.51QIKK69 pKa = 8.41YY70 pKa = 9.44EE71 pKa = 4.05KK72 pKa = 9.47KK73 pKa = 9.82QNHH76 pKa = 5.43TFIKK80 pKa = 10.17PKK82 pKa = 10.19LNLVIKK88 pKa = 9.65EE89 pKa = 4.14EE90 pKa = 3.78QHH92 pKa = 4.67ITKK95 pKa = 10.19KK96 pKa = 9.93VLRR99 pKa = 11.84GKK101 pKa = 9.79EE102 pKa = 3.64RR103 pKa = 11.84AATHH107 pKa = 6.45AFMKK111 pKa = 10.59EE112 pKa = 3.77MVEE115 pKa = 4.24SNKK118 pKa = 9.58IQPSWNVEE126 pKa = 3.87YY127 pKa = 10.38EE128 pKa = 4.09KK129 pKa = 11.01EE130 pKa = 3.82IDD132 pKa = 3.75EE133 pKa = 4.38VDD135 pKa = 3.57LFFMKK140 pKa = 10.47KK141 pKa = 7.05KK142 pKa = 8.73TKK144 pKa = 9.54PFSGFSIKK152 pKa = 8.72EE153 pKa = 3.95LRR155 pKa = 11.84DD156 pKa = 3.57SLIVQSDD163 pKa = 4.6DD164 pKa = 3.83KK165 pKa = 12.0NMAQPTVMSSIDD177 pKa = 3.79EE178 pKa = 4.08IVTPRR183 pKa = 11.84EE184 pKa = 4.17EE185 pKa = 4.07ISVSAISEE193 pKa = 4.05QLASLMEE200 pKa = 4.22RR201 pKa = 11.84VDD203 pKa = 4.13KK204 pKa = 11.07LEE206 pKa = 4.17KK207 pKa = 10.09MNAALEE213 pKa = 4.28EE214 pKa = 4.32EE215 pKa = 4.23NKK217 pKa = 9.9QLKK220 pKa = 9.77KK221 pKa = 10.5EE222 pKa = 3.94RR223 pKa = 11.84EE224 pKa = 4.06ATIKK228 pKa = 10.49SVKK231 pKa = 10.29KK232 pKa = 8.67EE233 pKa = 3.78AKK235 pKa = 9.75KK236 pKa = 10.34IKK238 pKa = 9.05QEE240 pKa = 3.72KK241 pKa = 7.73PQIVKK246 pKa = 8.68KK247 pKa = 7.73TQHH250 pKa = 6.4KK251 pKa = 10.37SLGVNLKK258 pKa = 8.15ITKK261 pKa = 9.03TKK263 pKa = 10.75VVGQEE268 pKa = 3.49QCLEE272 pKa = 4.1IEE274 pKa = 4.36NTQHH278 pKa = 6.69KK279 pKa = 10.39KK280 pKa = 10.27FVEE283 pKa = 4.4KK284 pKa = 10.32PSMPLKK290 pKa = 10.16VSKK293 pKa = 10.76KK294 pKa = 6.04MTEE297 pKa = 3.86HH298 pKa = 5.76QLKK301 pKa = 8.55KK302 pKa = 9.28TIRR305 pKa = 11.84TWYY308 pKa = 9.89EE309 pKa = 3.28FDD311 pKa = 3.51PSKK314 pKa = 10.74LVQHH318 pKa = 5.85QKK320 pKa = 10.43EE321 pKa = 4.36VLNSVVTNTTFADD334 pKa = 3.72KK335 pKa = 10.81VRR337 pKa = 11.84EE338 pKa = 3.97TGIPKK343 pKa = 10.0QKK345 pKa = 9.63IRR347 pKa = 11.84YY348 pKa = 6.1VAKK351 pKa = 10.12PPAEE355 pKa = 4.37EE356 pKa = 3.73KK357 pKa = 10.65RR358 pKa = 11.84SIHH361 pKa = 6.45FYY363 pKa = 10.09GYY365 pKa = 9.89KK366 pKa = 10.05PKK368 pKa = 10.89GIPNKK373 pKa = 9.81VWWNWVTTGTAMDD386 pKa = 4.52AYY388 pKa = 9.51EE389 pKa = 4.06KK390 pKa = 10.71ADD392 pKa = 3.62RR393 pKa = 11.84YY394 pKa = 10.58LYY396 pKa = 10.37HH397 pKa = 5.51QFKK400 pKa = 10.5RR401 pKa = 11.84EE402 pKa = 3.58MMIYY406 pKa = 9.28RR407 pKa = 11.84NKK409 pKa = 8.55WVKK412 pKa = 9.94FSKK415 pKa = 10.41EE416 pKa = 3.69FNPYY420 pKa = 9.42LSKK423 pKa = 11.02PKK425 pKa = 9.88MVWEE429 pKa = 4.26EE430 pKa = 3.98NTWEE434 pKa = 4.07YY435 pKa = 10.99EE436 pKa = 4.19YY437 pKa = 10.27KK438 pKa = 10.38TDD440 pKa = 3.64VPYY443 pKa = 11.45NFILKK448 pKa = 8.63WRR450 pKa = 11.84QLVQTYY456 pKa = 9.99KK457 pKa = 10.82PNTPIQADD465 pKa = 3.79WYY467 pKa = 10.68KK468 pKa = 10.56ISQKK472 pKa = 7.53QQCC475 pKa = 3.79
MM1 pKa = 7.82INNQTNKK8 pKa = 10.17KK9 pKa = 9.21GPQLEE14 pKa = 4.1RR15 pKa = 11.84VHH17 pKa = 6.82FGSTQVVGKK26 pKa = 7.26STKK29 pKa = 9.43RR30 pKa = 11.84RR31 pKa = 11.84QRR33 pKa = 11.84GTKK36 pKa = 9.87LDD38 pKa = 2.94IEE40 pKa = 4.57YY41 pKa = 9.64TVRR44 pKa = 11.84RR45 pKa = 11.84NDD47 pKa = 3.13APKK50 pKa = 9.57EE51 pKa = 3.95QKK53 pKa = 10.53FLISEE58 pKa = 4.1IFDD61 pKa = 3.51EE62 pKa = 5.62KK63 pKa = 10.92LDD65 pKa = 3.78KK66 pKa = 10.51QIKK69 pKa = 8.41YY70 pKa = 9.44EE71 pKa = 4.05KK72 pKa = 9.47KK73 pKa = 9.82QNHH76 pKa = 5.43TFIKK80 pKa = 10.17PKK82 pKa = 10.19LNLVIKK88 pKa = 9.65EE89 pKa = 4.14EE90 pKa = 3.78QHH92 pKa = 4.67ITKK95 pKa = 10.19KK96 pKa = 9.93VLRR99 pKa = 11.84GKK101 pKa = 9.79EE102 pKa = 3.64RR103 pKa = 11.84AATHH107 pKa = 6.45AFMKK111 pKa = 10.59EE112 pKa = 3.77MVEE115 pKa = 4.24SNKK118 pKa = 9.58IQPSWNVEE126 pKa = 3.87YY127 pKa = 10.38EE128 pKa = 4.09KK129 pKa = 11.01EE130 pKa = 3.82IDD132 pKa = 3.75EE133 pKa = 4.38VDD135 pKa = 3.57LFFMKK140 pKa = 10.47KK141 pKa = 7.05KK142 pKa = 8.73TKK144 pKa = 9.54PFSGFSIKK152 pKa = 8.72EE153 pKa = 3.95LRR155 pKa = 11.84DD156 pKa = 3.57SLIVQSDD163 pKa = 4.6DD164 pKa = 3.83KK165 pKa = 12.0NMAQPTVMSSIDD177 pKa = 3.79EE178 pKa = 4.08IVTPRR183 pKa = 11.84EE184 pKa = 4.17EE185 pKa = 4.07ISVSAISEE193 pKa = 4.05QLASLMEE200 pKa = 4.22RR201 pKa = 11.84VDD203 pKa = 4.13KK204 pKa = 11.07LEE206 pKa = 4.17KK207 pKa = 10.09MNAALEE213 pKa = 4.28EE214 pKa = 4.32EE215 pKa = 4.23NKK217 pKa = 9.9QLKK220 pKa = 9.77KK221 pKa = 10.5EE222 pKa = 3.94RR223 pKa = 11.84EE224 pKa = 4.06ATIKK228 pKa = 10.49SVKK231 pKa = 10.29KK232 pKa = 8.67EE233 pKa = 3.78AKK235 pKa = 9.75KK236 pKa = 10.34IKK238 pKa = 9.05QEE240 pKa = 3.72KK241 pKa = 7.73PQIVKK246 pKa = 8.68KK247 pKa = 7.73TQHH250 pKa = 6.4KK251 pKa = 10.37SLGVNLKK258 pKa = 8.15ITKK261 pKa = 9.03TKK263 pKa = 10.75VVGQEE268 pKa = 3.49QCLEE272 pKa = 4.1IEE274 pKa = 4.36NTQHH278 pKa = 6.69KK279 pKa = 10.39KK280 pKa = 10.27FVEE283 pKa = 4.4KK284 pKa = 10.32PSMPLKK290 pKa = 10.16VSKK293 pKa = 10.76KK294 pKa = 6.04MTEE297 pKa = 3.86HH298 pKa = 5.76QLKK301 pKa = 8.55KK302 pKa = 9.28TIRR305 pKa = 11.84TWYY308 pKa = 9.89EE309 pKa = 3.28FDD311 pKa = 3.51PSKK314 pKa = 10.74LVQHH318 pKa = 5.85QKK320 pKa = 10.43EE321 pKa = 4.36VLNSVVTNTTFADD334 pKa = 3.72KK335 pKa = 10.81VRR337 pKa = 11.84EE338 pKa = 3.97TGIPKK343 pKa = 10.0QKK345 pKa = 9.63IRR347 pKa = 11.84YY348 pKa = 6.1VAKK351 pKa = 10.12PPAEE355 pKa = 4.37EE356 pKa = 3.73KK357 pKa = 10.65RR358 pKa = 11.84SIHH361 pKa = 6.45FYY363 pKa = 10.09GYY365 pKa = 9.89KK366 pKa = 10.05PKK368 pKa = 10.89GIPNKK373 pKa = 9.81VWWNWVTTGTAMDD386 pKa = 4.52AYY388 pKa = 9.51EE389 pKa = 4.06KK390 pKa = 10.71ADD392 pKa = 3.62RR393 pKa = 11.84YY394 pKa = 10.58LYY396 pKa = 10.37HH397 pKa = 5.51QFKK400 pKa = 10.5RR401 pKa = 11.84EE402 pKa = 3.58MMIYY406 pKa = 9.28RR407 pKa = 11.84NKK409 pKa = 8.55WVKK412 pKa = 9.94FSKK415 pKa = 10.41EE416 pKa = 3.69FNPYY420 pKa = 9.42LSKK423 pKa = 11.02PKK425 pKa = 9.88MVWEE429 pKa = 4.26EE430 pKa = 3.98NTWEE434 pKa = 4.07YY435 pKa = 10.99EE436 pKa = 4.19YY437 pKa = 10.27KK438 pKa = 10.38TDD440 pKa = 3.64VPYY443 pKa = 11.45NFILKK448 pKa = 8.63WRR450 pKa = 11.84QLVQTYY456 pKa = 9.99KK457 pKa = 10.82PNTPIQADD465 pKa = 3.79WYY467 pKa = 10.68KK468 pKa = 10.56ISQKK472 pKa = 7.53QQCC475 pKa = 3.79
Molecular weight: 56.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3799 |
289 |
2104 |
949.8 |
107.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.87 ± 0.585 | 0.921 ± 0.296 |
5.238 ± 0.494 | 6.844 ± 0.623 |
3.79 ± 0.123 | 5.265 ± 0.712 |
1.764 ± 0.158 | 7.318 ± 0.357 |
7.37 ± 1.88 | 8.66 ± 0.902 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.053 ± 0.209 | 5.791 ± 0.353 |
4.291 ± 0.876 | 4.396 ± 0.493 |
4.528 ± 0.233 | 6.238 ± 0.265 |
7.37 ± 1.046 | 7.476 ± 0.636 |
1.474 ± 0.257 | 3.343 ± 0.116 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
