
Wuchan romanomermis nematode virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
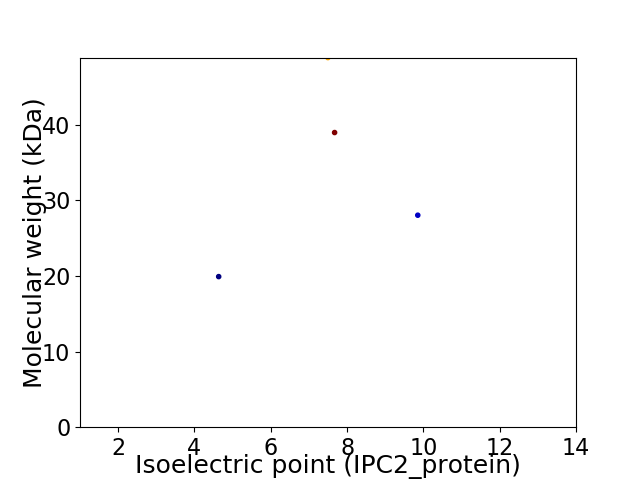
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KEY7|A0A1L3KEY7_9VIRU Uncharacterized protein OS=Wuchan romanomermis nematode virus 3 OX=1923687 PE=4 SV=1
MM1 pKa = 7.27WWDD4 pKa = 4.55FYY6 pKa = 9.39TWKK9 pKa = 10.35PYY11 pKa = 9.94IYY13 pKa = 10.37SVAGFNFTARR23 pKa = 11.84APQGLIPGEE32 pKa = 4.44LYY34 pKa = 9.31WLTVRR39 pKa = 11.84VVGPFLDD46 pKa = 3.41QFPFQPALSFSGFEE60 pKa = 3.94LVATLFSAIDD70 pKa = 3.41QLYY73 pKa = 8.85SAGVNVLTRR82 pKa = 11.84WVTKK86 pKa = 9.83IVVRR90 pKa = 11.84HH91 pKa = 4.34VTVDD95 pKa = 3.2PSYY98 pKa = 11.54SVDD101 pKa = 3.36PEE103 pKa = 3.69ILVRR107 pKa = 11.84FGQRR111 pKa = 11.84HH112 pKa = 4.17VDD114 pKa = 3.15IFAAAVRR121 pKa = 11.84DD122 pKa = 3.56YY123 pKa = 11.55ARR125 pKa = 11.84IYY127 pKa = 10.33ISMVFDD133 pKa = 3.68SLSGYY138 pKa = 8.87WPDD141 pKa = 4.1VPNQVDD147 pKa = 3.6ANSEE151 pKa = 4.26RR152 pKa = 11.84IGIPRR157 pKa = 11.84PMGDD161 pKa = 3.32GADD164 pKa = 3.64SLEE167 pKa = 4.39TSWEE171 pKa = 4.3VVSEE175 pKa = 4.33GG176 pKa = 4.04
MM1 pKa = 7.27WWDD4 pKa = 4.55FYY6 pKa = 9.39TWKK9 pKa = 10.35PYY11 pKa = 9.94IYY13 pKa = 10.37SVAGFNFTARR23 pKa = 11.84APQGLIPGEE32 pKa = 4.44LYY34 pKa = 9.31WLTVRR39 pKa = 11.84VVGPFLDD46 pKa = 3.41QFPFQPALSFSGFEE60 pKa = 3.94LVATLFSAIDD70 pKa = 3.41QLYY73 pKa = 8.85SAGVNVLTRR82 pKa = 11.84WVTKK86 pKa = 9.83IVVRR90 pKa = 11.84HH91 pKa = 4.34VTVDD95 pKa = 3.2PSYY98 pKa = 11.54SVDD101 pKa = 3.36PEE103 pKa = 3.69ILVRR107 pKa = 11.84FGQRR111 pKa = 11.84HH112 pKa = 4.17VDD114 pKa = 3.15IFAAAVRR121 pKa = 11.84DD122 pKa = 3.56YY123 pKa = 11.55ARR125 pKa = 11.84IYY127 pKa = 10.33ISMVFDD133 pKa = 3.68SLSGYY138 pKa = 8.87WPDD141 pKa = 4.1VPNQVDD147 pKa = 3.6ANSEE151 pKa = 4.26RR152 pKa = 11.84IGIPRR157 pKa = 11.84PMGDD161 pKa = 3.32GADD164 pKa = 3.64SLEE167 pKa = 4.39TSWEE171 pKa = 4.3VVSEE175 pKa = 4.33GG176 pKa = 4.04
Molecular weight: 19.93 kDa
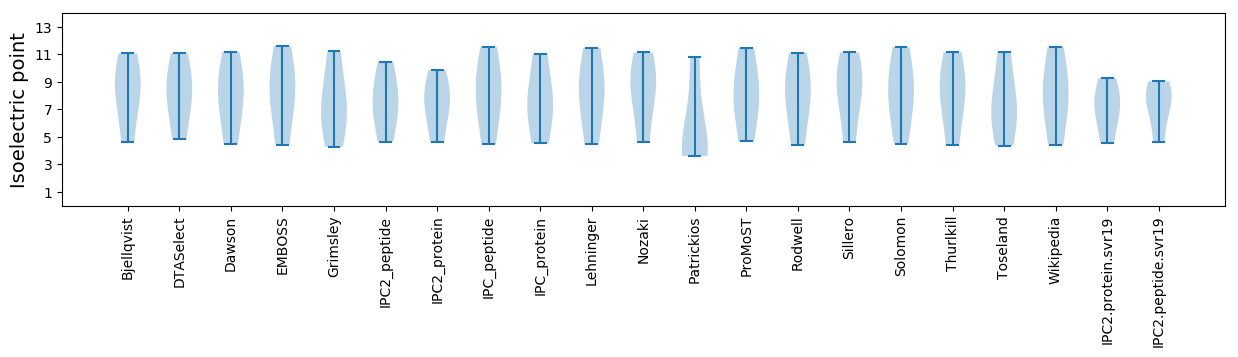
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEX8|A0A1L3KEX8_9VIRU Uncharacterized protein OS=Wuchan romanomermis nematode virus 3 OX=1923687 PE=4 SV=1
MM1 pKa = 7.11LQAPVRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84QNNNGGLIGQLVRR26 pKa = 11.84QLASLEE32 pKa = 4.15LARR35 pKa = 11.84APAVQQPQPARR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84GAGGAGGLITAGPSATVATRR74 pKa = 11.84GGPSFANSPQGGLRR88 pKa = 11.84LVNEE92 pKa = 4.79EE93 pKa = 4.03LWKK96 pKa = 11.09SADD99 pKa = 2.92ITTADD104 pKa = 4.22FLQKK108 pKa = 10.04LTFTPGKK115 pKa = 10.04SGLTILDD122 pKa = 4.53RR123 pKa = 11.84IAATFDD129 pKa = 3.29RR130 pKa = 11.84YY131 pKa = 10.07VVHH134 pKa = 6.67SVSLSWRR141 pKa = 11.84TSVGTQKK148 pKa = 10.87DD149 pKa = 3.36GAYY152 pKa = 10.46VIGIEE157 pKa = 3.89WDD159 pKa = 3.42ASKK162 pKa = 9.03TAPNYY167 pKa = 11.0ADD169 pKa = 3.48AQALVPRR176 pKa = 11.84IRR178 pKa = 11.84SAIWKK183 pKa = 8.31EE184 pKa = 3.82EE185 pKa = 3.74QMVLPSEE192 pKa = 4.33RR193 pKa = 11.84LMSRR197 pKa = 11.84RR198 pKa = 11.84FYY200 pKa = 10.86STGFDD205 pKa = 3.86DD206 pKa = 5.97AATKK210 pKa = 11.01NPDD213 pKa = 3.21CTAFAAILAVKK224 pKa = 10.02GVASAGNVGDD234 pKa = 3.04VWIRR238 pKa = 11.84YY239 pKa = 8.43DD240 pKa = 3.41ISLMGPTSANSKK252 pKa = 8.17TAVLEE257 pKa = 4.11NN258 pKa = 3.76
MM1 pKa = 7.11LQAPVRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84QNNNGGLIGQLVRR26 pKa = 11.84QLASLEE32 pKa = 4.15LARR35 pKa = 11.84APAVQQPQPARR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84GAGGAGGLITAGPSATVATRR74 pKa = 11.84GGPSFANSPQGGLRR88 pKa = 11.84LVNEE92 pKa = 4.79EE93 pKa = 4.03LWKK96 pKa = 11.09SADD99 pKa = 2.92ITTADD104 pKa = 4.22FLQKK108 pKa = 10.04LTFTPGKK115 pKa = 10.04SGLTILDD122 pKa = 4.53RR123 pKa = 11.84IAATFDD129 pKa = 3.29RR130 pKa = 11.84YY131 pKa = 10.07VVHH134 pKa = 6.67SVSLSWRR141 pKa = 11.84TSVGTQKK148 pKa = 10.87DD149 pKa = 3.36GAYY152 pKa = 10.46VIGIEE157 pKa = 3.89WDD159 pKa = 3.42ASKK162 pKa = 9.03TAPNYY167 pKa = 11.0ADD169 pKa = 3.48AQALVPRR176 pKa = 11.84IRR178 pKa = 11.84SAIWKK183 pKa = 8.31EE184 pKa = 3.82EE185 pKa = 3.74QMVLPSEE192 pKa = 4.33RR193 pKa = 11.84LMSRR197 pKa = 11.84RR198 pKa = 11.84FYY200 pKa = 10.86STGFDD205 pKa = 3.86DD206 pKa = 5.97AATKK210 pKa = 11.01NPDD213 pKa = 3.21CTAFAAILAVKK224 pKa = 10.02GVASAGNVGDD234 pKa = 3.04VWIRR238 pKa = 11.84YY239 pKa = 8.43DD240 pKa = 3.41ISLMGPTSANSKK252 pKa = 8.17TAVLEE257 pKa = 4.11NN258 pKa = 3.76
Molecular weight: 28.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1218 |
176 |
448 |
304.5 |
33.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.031 ± 0.965 | 1.149 ± 0.374 |
5.747 ± 0.398 | 4.516 ± 0.623 |
4.023 ± 0.873 | 7.882 ± 0.816 |
1.478 ± 0.308 | 4.598 ± 0.33 |
3.777 ± 0.554 | 9.113 ± 0.568 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.135 ± 0.364 | 2.709 ± 0.455 |
5.255 ± 0.302 | 4.433 ± 0.248 |
7.964 ± 0.832 | 7.882 ± 0.414 |
4.351 ± 0.567 | 7.964 ± 0.917 |
2.545 ± 0.681 | 3.448 ± 0.493 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
