
Porcine polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; unclassified Polyomaviridae
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
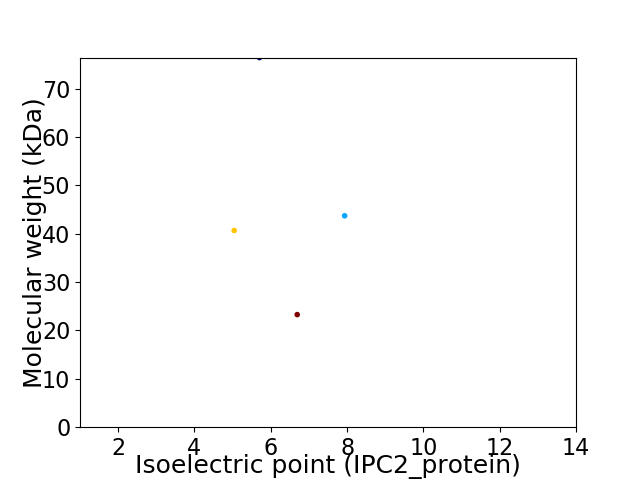
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1K4R5|A0A2P1K4R5_9POLY VP2 OS=Porcine polyomavirus OX=2126099 PE=4 SV=1
MM1 pKa = 7.29GAILAIPEE9 pKa = 3.83ILAAAVEE16 pKa = 4.65GAAEE20 pKa = 4.68AIEE23 pKa = 4.13IAGSFGAVASGEE35 pKa = 3.94GLAAVEE41 pKa = 4.4TLVQSATLGGEE52 pKa = 3.99YY53 pKa = 10.24LGASLAEE60 pKa = 3.83QAAALGIPDD69 pKa = 3.84SAFTLLSQTPEE80 pKa = 3.76LANTFTAISGLVNAGAGLAGGVATYY105 pKa = 10.44LGNSGGLPGQEE116 pKa = 3.91AGGGGSGLYY125 pKa = 9.61PYY127 pKa = 10.81YY128 pKa = 9.47PGHH131 pKa = 6.83GGHH134 pKa = 6.48FHH136 pKa = 6.63PTHH139 pKa = 6.13HH140 pKa = 6.1TMEE143 pKa = 4.72LAVIPPYY150 pKa = 10.23QLEE153 pKa = 4.18SGIPGIPDD161 pKa = 3.0WVLNLLPEE169 pKa = 4.63LPSLSDD175 pKa = 3.19IVSGIANGLWTSYY188 pKa = 10.65YY189 pKa = 9.26RR190 pKa = 11.84TGTEE194 pKa = 4.47LIRR197 pKa = 11.84RR198 pKa = 11.84TASDD202 pKa = 3.32EE203 pKa = 4.03LQRR206 pKa = 11.84LLGDD210 pKa = 4.07LSTGLRR216 pKa = 11.84QASEE220 pKa = 3.93AAQQYY225 pKa = 9.51IQDD228 pKa = 4.3ADD230 pKa = 3.73PVNALVRR237 pKa = 11.84QIEE240 pKa = 4.2RR241 pKa = 11.84VQVQRR246 pKa = 11.84RR247 pKa = 11.84VDD249 pKa = 3.54SARR252 pKa = 11.84LIQEE256 pKa = 4.17AEE258 pKa = 4.09TGVLNLGEE266 pKa = 4.07IVQRR270 pKa = 11.84VADD273 pKa = 4.21TTRR276 pKa = 11.84GAAGSVGQIATDD288 pKa = 4.43LAQLPVDD295 pKa = 4.96GYY297 pKa = 10.1NALSGGVQRR306 pKa = 11.84LGQWISMSGPTGGTLHH322 pKa = 6.68YY323 pKa = 10.54SFPHH327 pKa = 4.91WVLYY331 pKa = 10.64VLDD334 pKa = 4.38EE335 pKa = 4.5LQLDD339 pKa = 4.45FPPKK343 pKa = 9.87QGIKK347 pKa = 10.1RR348 pKa = 11.84GLPEE352 pKa = 5.31DD353 pKa = 4.01PQPPEE358 pKa = 3.9GDD360 pKa = 3.3VYY362 pKa = 10.24QEE364 pKa = 3.54RR365 pKa = 11.84KK366 pKa = 7.53TRR368 pKa = 11.84LRR370 pKa = 11.84TYY372 pKa = 10.89KK373 pKa = 10.47SGPKK377 pKa = 8.29TANPNKK383 pKa = 9.64KK384 pKa = 9.53RR385 pKa = 11.84RR386 pKa = 11.84RR387 pKa = 3.69
MM1 pKa = 7.29GAILAIPEE9 pKa = 3.83ILAAAVEE16 pKa = 4.65GAAEE20 pKa = 4.68AIEE23 pKa = 4.13IAGSFGAVASGEE35 pKa = 3.94GLAAVEE41 pKa = 4.4TLVQSATLGGEE52 pKa = 3.99YY53 pKa = 10.24LGASLAEE60 pKa = 3.83QAAALGIPDD69 pKa = 3.84SAFTLLSQTPEE80 pKa = 3.76LANTFTAISGLVNAGAGLAGGVATYY105 pKa = 10.44LGNSGGLPGQEE116 pKa = 3.91AGGGGSGLYY125 pKa = 9.61PYY127 pKa = 10.81YY128 pKa = 9.47PGHH131 pKa = 6.83GGHH134 pKa = 6.48FHH136 pKa = 6.63PTHH139 pKa = 6.13HH140 pKa = 6.1TMEE143 pKa = 4.72LAVIPPYY150 pKa = 10.23QLEE153 pKa = 4.18SGIPGIPDD161 pKa = 3.0WVLNLLPEE169 pKa = 4.63LPSLSDD175 pKa = 3.19IVSGIANGLWTSYY188 pKa = 10.65YY189 pKa = 9.26RR190 pKa = 11.84TGTEE194 pKa = 4.47LIRR197 pKa = 11.84RR198 pKa = 11.84TASDD202 pKa = 3.32EE203 pKa = 4.03LQRR206 pKa = 11.84LLGDD210 pKa = 4.07LSTGLRR216 pKa = 11.84QASEE220 pKa = 3.93AAQQYY225 pKa = 9.51IQDD228 pKa = 4.3ADD230 pKa = 3.73PVNALVRR237 pKa = 11.84QIEE240 pKa = 4.2RR241 pKa = 11.84VQVQRR246 pKa = 11.84RR247 pKa = 11.84VDD249 pKa = 3.54SARR252 pKa = 11.84LIQEE256 pKa = 4.17AEE258 pKa = 4.09TGVLNLGEE266 pKa = 4.07IVQRR270 pKa = 11.84VADD273 pKa = 4.21TTRR276 pKa = 11.84GAAGSVGQIATDD288 pKa = 4.43LAQLPVDD295 pKa = 4.96GYY297 pKa = 10.1NALSGGVQRR306 pKa = 11.84LGQWISMSGPTGGTLHH322 pKa = 6.68YY323 pKa = 10.54SFPHH327 pKa = 4.91WVLYY331 pKa = 10.64VLDD334 pKa = 4.38EE335 pKa = 4.5LQLDD339 pKa = 4.45FPPKK343 pKa = 9.87QGIKK347 pKa = 10.1RR348 pKa = 11.84GLPEE352 pKa = 5.31DD353 pKa = 4.01PQPPEE358 pKa = 3.9GDD360 pKa = 3.3VYY362 pKa = 10.24QEE364 pKa = 3.54RR365 pKa = 11.84KK366 pKa = 7.53TRR368 pKa = 11.84LRR370 pKa = 11.84TYY372 pKa = 10.89KK373 pKa = 10.47SGPKK377 pKa = 8.29TANPNKK383 pKa = 9.64KK384 pKa = 9.53RR385 pKa = 11.84RR386 pKa = 11.84RR387 pKa = 3.69
Molecular weight: 40.68 kDa
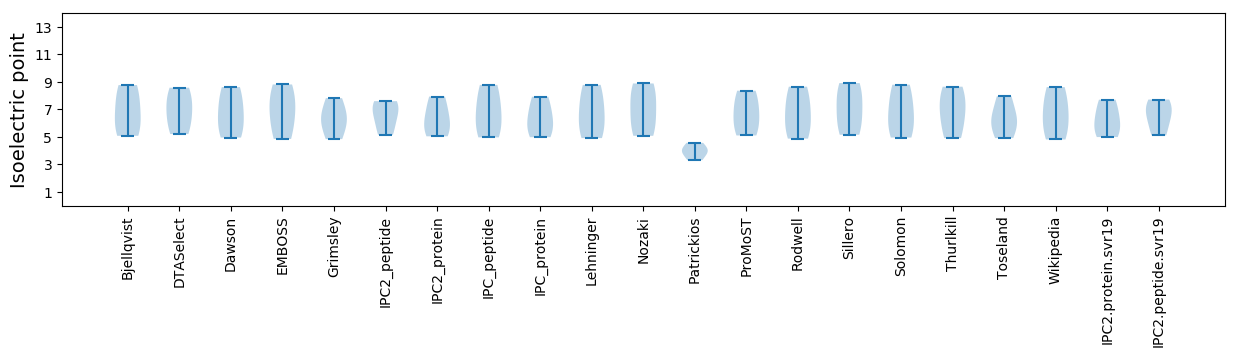
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1K4Q5|A0A2P1K4Q5_9POLY Large T antigen OS=Porcine polyomavirus OX=2126099 PE=4 SV=1
MM1 pKa = 7.47SWMNFSLISPQNRR14 pKa = 11.84ASRR17 pKa = 11.84EE18 pKa = 3.76ACLRR22 pKa = 11.84IHH24 pKa = 6.07SHH26 pKa = 6.47LKK28 pKa = 10.49GMSTKK33 pKa = 10.1KK34 pKa = 10.26GKK36 pKa = 9.97RR37 pKa = 11.84VSAPTRR43 pKa = 11.84VVQRR47 pKa = 11.84LPTQIRR53 pKa = 11.84KK54 pKa = 10.16GGVDD58 pKa = 3.39VLATVPLDD66 pKa = 3.04QHH68 pKa = 5.56TEE70 pKa = 4.05YY71 pKa = 10.93KK72 pKa = 10.51VEE74 pKa = 4.42LFLKK78 pKa = 8.97PTFATTDD85 pKa = 3.39GNGVFQSIGGPLTNGQEE102 pKa = 4.24PNVGEE107 pKa = 4.14LLLYY111 pKa = 10.22SLGMVQPPEE120 pKa = 3.94IPNQVDD126 pKa = 3.87EE127 pKa = 4.62NSMIVWEE134 pKa = 4.62LYY136 pKa = 9.8RR137 pKa = 11.84VEE139 pKa = 4.34TEE141 pKa = 4.02VLFAPKK147 pKa = 9.58TFSTGYY153 pKa = 9.45TSAQKK158 pKa = 10.85NIGGVEE164 pKa = 4.23GPQFYY169 pKa = 9.69FWAVGGEE176 pKa = 4.0PLDD179 pKa = 3.56VLGIKK184 pKa = 9.15SRR186 pKa = 11.84KK187 pKa = 8.69SVTYY191 pKa = 7.44PTGVQAPPNGEE202 pKa = 4.1DD203 pKa = 3.5LVTEE207 pKa = 3.86NTVRR211 pKa = 11.84RR212 pKa = 11.84KK213 pKa = 9.39VDD215 pKa = 3.38KK216 pKa = 10.46PQFPCDD222 pKa = 3.3CWLPDD227 pKa = 4.06PSRR230 pKa = 11.84NDD232 pKa = 2.88NTRR235 pKa = 11.84YY236 pKa = 8.56FGRR239 pKa = 11.84VIGGSQTPPVLTIGNSSTVPVVDD262 pKa = 4.22EE263 pKa = 4.46YY264 pKa = 11.84GVGVLLLQGRR274 pKa = 11.84LYY276 pKa = 9.84VTSCDD281 pKa = 3.53MLGTVAEE288 pKa = 4.42VNEE291 pKa = 4.13ATLAPEE297 pKa = 4.2SNSRR301 pKa = 11.84RR302 pKa = 11.84IHH304 pKa = 5.9AGSGRR309 pKa = 11.84FFRR312 pKa = 11.84LHH314 pKa = 4.93FRR316 pKa = 11.84QRR318 pKa = 11.84KK319 pKa = 7.63VKK321 pKa = 10.47NPYY324 pKa = 9.06TINLLYY330 pKa = 10.54KK331 pKa = 10.35QVFNKK336 pKa = 10.42PNDD339 pKa = 3.71DD340 pKa = 3.71FVGQTGVTEE349 pKa = 3.94VTMTQEE355 pKa = 4.24TQPLPTILEE364 pKa = 4.48GTVGTQGNLVTEE376 pKa = 4.46ALRR379 pKa = 11.84RR380 pKa = 11.84GTAVLQTTTPLVNTQSLL397 pKa = 3.7
MM1 pKa = 7.47SWMNFSLISPQNRR14 pKa = 11.84ASRR17 pKa = 11.84EE18 pKa = 3.76ACLRR22 pKa = 11.84IHH24 pKa = 6.07SHH26 pKa = 6.47LKK28 pKa = 10.49GMSTKK33 pKa = 10.1KK34 pKa = 10.26GKK36 pKa = 9.97RR37 pKa = 11.84VSAPTRR43 pKa = 11.84VVQRR47 pKa = 11.84LPTQIRR53 pKa = 11.84KK54 pKa = 10.16GGVDD58 pKa = 3.39VLATVPLDD66 pKa = 3.04QHH68 pKa = 5.56TEE70 pKa = 4.05YY71 pKa = 10.93KK72 pKa = 10.51VEE74 pKa = 4.42LFLKK78 pKa = 8.97PTFATTDD85 pKa = 3.39GNGVFQSIGGPLTNGQEE102 pKa = 4.24PNVGEE107 pKa = 4.14LLLYY111 pKa = 10.22SLGMVQPPEE120 pKa = 3.94IPNQVDD126 pKa = 3.87EE127 pKa = 4.62NSMIVWEE134 pKa = 4.62LYY136 pKa = 9.8RR137 pKa = 11.84VEE139 pKa = 4.34TEE141 pKa = 4.02VLFAPKK147 pKa = 9.58TFSTGYY153 pKa = 9.45TSAQKK158 pKa = 10.85NIGGVEE164 pKa = 4.23GPQFYY169 pKa = 9.69FWAVGGEE176 pKa = 4.0PLDD179 pKa = 3.56VLGIKK184 pKa = 9.15SRR186 pKa = 11.84KK187 pKa = 8.69SVTYY191 pKa = 7.44PTGVQAPPNGEE202 pKa = 4.1DD203 pKa = 3.5LVTEE207 pKa = 3.86NTVRR211 pKa = 11.84RR212 pKa = 11.84KK213 pKa = 9.39VDD215 pKa = 3.38KK216 pKa = 10.46PQFPCDD222 pKa = 3.3CWLPDD227 pKa = 4.06PSRR230 pKa = 11.84NDD232 pKa = 2.88NTRR235 pKa = 11.84YY236 pKa = 8.56FGRR239 pKa = 11.84VIGGSQTPPVLTIGNSSTVPVVDD262 pKa = 4.22EE263 pKa = 4.46YY264 pKa = 11.84GVGVLLLQGRR274 pKa = 11.84LYY276 pKa = 9.84VTSCDD281 pKa = 3.53MLGTVAEE288 pKa = 4.42VNEE291 pKa = 4.13ATLAPEE297 pKa = 4.2SNSRR301 pKa = 11.84RR302 pKa = 11.84IHH304 pKa = 5.9AGSGRR309 pKa = 11.84FFRR312 pKa = 11.84LHH314 pKa = 4.93FRR316 pKa = 11.84QRR318 pKa = 11.84KK319 pKa = 7.63VKK321 pKa = 10.47NPYY324 pKa = 9.06TINLLYY330 pKa = 10.54KK331 pKa = 10.35QVFNKK336 pKa = 10.42PNDD339 pKa = 3.71DD340 pKa = 3.71FVGQTGVTEE349 pKa = 3.94VTMTQEE355 pKa = 4.24TQPLPTILEE364 pKa = 4.48GTVGTQGNLVTEE376 pKa = 4.46ALRR379 pKa = 11.84RR380 pKa = 11.84GTAVLQTTTPLVNTQSLL397 pKa = 3.7
Molecular weight: 43.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1634 |
195 |
655 |
408.5 |
46.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.263 ± 1.62 | 2.142 ± 0.989 |
5.141 ± 0.805 | 6.548 ± 0.396 |
3.55 ± 0.743 | 8.078 ± 1.673 |
2.02 ± 0.314 | 4.162 ± 0.307 |
5.447 ± 1.291 | 10.71 ± 0.562 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.326 ± 0.563 | 4.651 ± 0.657 |
5.202 ± 0.939 | 4.835 ± 0.51 |
5.263 ± 0.278 | 5.447 ± 0.332 |
6.426 ± 1.132 | 6.977 ± 1.064 |
1.836 ± 0.67 | 3.978 ± 0.548 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
